Linking COPASI and Cytoscape
Hikmet Emre Kaya
Hello Everyone!
For my PhD project, I need to link COPASI and Cytoscape.
Cytoscape is an open source bioinformatics software platform for visualizing molecular interaction networks and integrating with gene expression profiles and other state data.
It has an app store where you can install apps for additional features, but there is no app for what I want to do, which is to run reaction kinetics simulations on the networks, particularly glycosylation networks.
So, I want to make COPASI available and compatible with Cytoscape.
Because Cytoscape is a Java application, I downloaded the java bindings for COPASI and decompiled the .class files to .java files. But, I am not sure how to proceed from here. Using Eclipse, I created a new maven project in the Cytoscape-api repository and added the .java files for COPASI.
I think I need to update the pom.xml file to add the COPASI dependencies, but I am not sure how to do that.
Sorry if the question sounds too vague, I guess I need an overall idea of how to best go about this.
Thanks,
Emre
Frank Bergmann
Hikmet Emre Kaya
At the moment, when I copy the .jar file to CytoscapeConfiguration/3/apps/installed, Cytoscape crashes and gives the following error:
Native code library failed to load.
java.lang.UnsatisfiedLinkError: no CopasiJava in java.library.path: [/home/people/hkaya/Cytoscape_v3.8.2/framework/lib, /home/people/hkaya/Cytoscape_v3.8.2/framework/lib, /usr/java/packages/lib, /usr/lib64, /lib64, /lib, /usr/lib]
Frank Bergmann
Hikmet Emre Kaya
That makes sense; I do work on linux x64.
I will try this and keep you posted about the outcome. Thanks!
Hikmet Emre Kaya
So, linking the native library to my java.library.path did work, but COPASI still fails to start when I open Cytoscape and gives the following error:
org.cytoscape.app.internal.exception.AppStartupException: Bundle start error
at org.cytoscape.app.internal.manager.BundleApp.start(BundleApp.java:84) ~[?:?]
at org.cytoscape.app.internal.manager.AppManager.startApps(AppManager.java:404) ~[?:?]
at org.cytoscape.app.internal.manager.AppManager.initializeApps(AppManager.java:374) ~[?:?]
at org.cytoscape.app.internal.manager.AppManager.lambda$attemptInitialization$0(AppManager.java:235) ~[?:?]
at java.util.concurrent.Executors$RunnableAdapter.call(Executors.java:515) [?:?]
at java.util.concurrent.FutureTask.run(FutureTask.java:264) [?:?]
at java.util.concurrent.ThreadPoolExecutor.runWorker(ThreadPoolExecutor.java:1128) [?:?]
at java.util.concurrent.ThreadPoolExecutor$Worker.run(ThreadPoolExecutor.java:628) [?:?]
at java.lang.Thread.run(Thread.java:834) [?:?]
Caused by: org.osgi.framework.BundleException: Activator start error in bundle org.cytoscape.COPASI [123].
at org.apache.felix.framework.Felix.activateBundle(Felix.java:2290) ~[?:?]
at org.apache.felix.framework.Felix.startBundle(Felix.java:2146) ~[?:?]
at org.apache.felix.framework.BundleImpl.start(BundleImpl.java:998) ~[?:?]
at org.apache.felix.framework.BundleImpl.start(BundleImpl.java:984) ~[?:?]
at org.cytoscape.app.internal.manager.BundleApp.start(BundleApp.java:82) ~[?:?]
... 8 more
Caused by: java.lang.UnsatisfiedLinkError: 'void org.cytoscape.COPASI.COPASIJNI.initCopasi()'
at org.cytoscape.COPASI.COPASIJNI.initCopasi(Native Method) ~[?:?]
at org.cytoscape.COPASI.COPASIJNI.<clinit>(COPASIJNI.java:25) ~[?:?]
at org.cytoscape.COPASI.CCopasiMessage.<init>(CCopasiMessage.java:87) ~[?:?]
at org.cytoscape.COPASI.CyActivator.start(CyActivator.java:52) ~[?:?]
at org.apache.felix.framework.util.SecureAction.startActivator(SecureAction.java:697) ~[?:?]
at org.apache.felix.framework.Felix.activateBundle(Felix.java:2240) ~[?:?]
at org.apache.felix.framework.Felix.startBundle(Felix.java:2146) ~[?:?]
at org.apache.felix.framework.BundleImpl.start(BundleImpl.java:998) ~[?:?]
at org.apache.felix.framework.BundleImpl.start(BundleImpl.java:984) ~[?:?]
at org.cytoscape.app.internal.manager.BundleApp.start(BundleApp.java:82) ~[?:?]
... 8 more
So, I probably need to define the COPASI library dependencies in my pom.xml file. I followed the instructions here to compile the dependencies, but it didn't solve the problem. How can these dependencies be installed locally using maven?
Frank Bergmann
at org.cytoscape.COPASI.COPASIJNI.initCopasi(Native Method) ~[?:?]
Hikmet Emre Kaya
Frank Bergmann
Hikmet Emre Kaya
I have managed to include the .so file in my jar, and the app starts without crashing, but does not appear on the menu tab in Cytoscape. I figured this might be because of the way I am activating it.
I want to clarify: when accessing the api of COPASI in the java project, does one have to do System.loadLibrary to load the native library? If this is already done in the c++ wrapper COPASIJNI.class, which many other COPASI classes depend on, do we still need to declare these classes as native in our source code?
Some examples contain System.loadLibrary and some don't, so I got confused
I hope my question makes sense.
Frank Bergmann
Hikmet Emre Kaya
I am not sure but I previously found the following links when investigating this issue. See if they are relevant?
https://github.com/LWJGL/lwjgl3/issues/277
https://stackoverflow.com/questions/11783632/how-do-i-load-and-use-native-library-in-java
I am trying to figure out if there is a way to call copasi.jar from my bundle app with a command. Because the jar doesn't have a main class, calling java -jar copasi.jar will return a "no main attribute..." error. So, I am not sure how to implement the main class and utilize this jar file as an executable. I understand that one can type java -cp copasi.jar mainclass.java (where mainclass.java can be one of the example files), but it is not really the best way to do it inside a bundle app, I suppose.
What do you think?
Frank Bergmann
Frank
Hikmet Emre Kaya
My initial idea was to add the following to the unimplemented public void run method at the end of Example1.java
ExecuteCommandTask executeCommandTask = new ExecuteCommandTask(command);
Where the command would be something like "java -jar copasi.jar" or "java -cp copasi.jar main.class" that would initialize COPASI
I hope I am making sense :D Calling myself a novice in app development would be an understatement :)
Frank Bergmann
Hikmet Emre Kaya
Hikmet Emre Kaya
Before asking my other questions, here is how it looks on my Linux machine.
As per the ReadMe instructions, I have mvn-installed copasi.jar file, edited my pom.xml and copied the .so file to /usr/lib/ Interestingly, it reads .xml files but gives an unsatisfiedLinkError when trying to open .cps files
java.lang.UnsatisfiedLinkError: 'boolean org.COPASI.COPASIJNI.CDataModel_loadFromString__SWIG_3(long, org.COPASI.CDataModel, java.lang.String)'
at org.COPASI.COPASIJNI.CDataModel_loadFromString__SWIG_3(Native Method) ~[?:?]
at org.COPASI.CDataModel.loadFromString(CDataModel.java:88) ~[?:?]
at org.copasi.cytoscape.internal.tasks.CopasiFileReaderTask.run(CopasiFileReaderTask.java:165) ~[?:?]
at org.cytoscape.work.internal.task.JDialogTaskManager$TaskRunnable.innerRun(JDialogTaskManager.java:337) ~[93:org.cytoscape.work-swing-impl:3.8.2]
at org.cytoscape.work.internal.task.JDialogTaskManager$TaskRunnable.run(JDialogTaskManager.java:352) [93:org.cytoscape.work-swing-impl:3.8.2]
at java.util.concurrent.Executors$RunnableAdapter.call(Executors.java:515) [?:?]
at java.util.concurrent.FutureTask.run(FutureTask.java:264) [?:?]
at java.util.concurrent.ThreadPoolExecutor.runWorker(ThreadPoolExecutor.java:1128) [?:?]
at java.util.concurrent.ThreadPoolExecutor$Worker.run(ThreadPoolExecutor.java:628) [?:?]
at java.lang.Thread.run(Thread.java:834) [?:?]
at org.cytoscape.work.internal.task.JDialogTaskManager$TaskRunnable.run(JDialogTaskManager.java:362)
at java.base/java.util.concurrent.Executors$RunnableAdapter.call(Executors.java:515)
at java.base/java.util.concurrent.FutureTask.run(FutureTask.java:264)
at java.base/java.util.concurrent.ThreadPoolExecutor.runWorker(ThreadPoolExecutor.java:1128)
at java.base/java.util.concurrent.ThreadPoolExecutor$Worker.run(ThreadPoolExecutor.java:628)
at java.base/java.lang.Thread.run(Thread.java:834)
Caused by: java.lang.UnsatisfiedLinkError: 'boolean org.COPASI.COPASIJNI.CDataModel_loadFromString__SWIG_3(long, org.COPASI.CDataModel, java.lang.String)'
at org.COPASI.COPASIJNI.CDataModel_loadFromString__SWIG_3(Native Method)
at org.COPASI.CDataModel.loadFromString(CDataModel.java:88)
at org.copasi.cytoscape.internal.tasks.CopasiFileReaderTask.run(CopasiFileReaderTask.java:165)
at org.cytoscape.work.internal.task.JDialogTaskManager$TaskRunnable.innerRun(JDialogTaskManager.java:337)
at org.cytoscape.work.internal.task.JDialogTaskManager$TaskRunnable.run(JDialogTaskManager.java:352)
Have you encountered this error when you tried it? Why would it specifically give the error with a .cps file, but not with an .xml file?
Frank Bergmann
Hikmet Emre Kaya
I definitely took the copasi.jar and libCopasiJava.so files from your repository, and then I copied the .so file to /usr/lib as a root (sudo cp ...). I didn't take the copasi-gui.jar file, though.
Hikmet Emre Kaya
Interestingly, when I typed export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/home/people/hkaya/git/cytoscape-api/CytoCopasi/lib/ (to add an accessible location to the java.library.path) manually, I was able to import a .cps file
Perhaps, I could try to use System.setProperty to add the location of the .so file to my java.library.path without having to copy the .so file itself.
Frank Bergmann
--
You received this message because you are subscribed to the Google Groups "COPASI User Forum" group.
To unsubscribe from this group and stop receiving emails from it, send an email to copasi-user-fo...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/copasi-user-forum/50ca5aea-fb18-4589-bfd4-588eb8371d23n%40googlegroups.com.
Hikmet Emre Kaya
I hope you have been doing well.
I put the native library issue aside later to deal with when it's time to think about app submission. In the meantime, I was able to expand the app's features so it can make conversion between .sbml and .cps files (i.e., save one as the other)
I am now wanting to create plots for time course and steady state simulations, but am confused about how to make use of the classes in copasi.jar . More specifically, does one first have to generate a report to create a plot? Or can I generate a plot directly?
I don't really understand what purpose PlotSpecification, CPlotItem , CPlotDataChannelSpec.java, etc each serve :/
Hikmet Emre Kaya
My SimulationTask class will briefly pop up a dialog (in a similar way to CellDesigner) to ask for duration, number of steps, etc, as well as the plot type (which I didn't have any problem writing)
Frank Bergmann
Hikmet Emre Kaya
Thank you for the suggestion. That actually worked perfectly! I used JFreeChart library to obtain plots.
My next question is; how would one go about getting flux rate vs time? I saw the getFlux() method, but the Copasi documentation says it only gives the final flux (which I tested and confirmed)
Thanks
Emre
Hikmet Emre Kaya
I have made quite a lot of progress with COPASI's implementation of CytoScape, and I managed to generate plots for Time-Course Simulations, as well as running Steady-State Calculations.
I am currently working on writing the Optimization Task. I created the user dialog to specify the objective function, the parameter, and the optimization method, but I got stuck at some point.
I am not sure how to create expressions for the objective function. For example, if the user wants to calculate the ratios of two reaction fluxes, how can one generate that expression? I can parse reaction names, but then the app would not understand that I am referring to their fluxes. Is there a method to specify those? I hope my question is clear enough.
Best,
Emre
Hikmet Emre Kaya
When you click on a reaction flux, the expression looks like this:
{(Adenylate kinase).Flux}
How does this work in COPASI's java bindings?
Frank Bergmann
You received this message because you are subscribed to a topic in the Google Groups "COPASI User Forum" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/copasi-user-forum/g_gVsfDofOE/unsubscribe.
To unsubscribe from this group and all its topics, send an email to copasi-user-fo...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/copasi-user-forum/5ed6a4b9-2bac-41e2-bd72-c09b1e62e32an%40googlegroups.com.
Hikmet Emre Kaya
Thank you for the detailed explanation. By that logic, I can get an expression for the flux by model.getReaction(index).getFluxReference().getCN.toString(); correct?
I just didn't quite understand the following:
Do you mean I should save the plot as a txt file to find the ChannelSpec?
Hikmet Emre Kaya
I can't figure out how to get the CN format for the reaction parameters. I can get parameter names from model.getReaction(index).getParameters.getName(), but I don't think that can be put into an expression.
Frank Bergmann
Frank Bergmann
Thank you for the detailed explanation. By that logic, I can get an expression for the flux by model.getReaction(index).getFluxReference().getCN.toString(); correct?
I just didn't quite understand the following:but I would suggest that you manually create a plot for the elements you want to use, open it in a text editor, and look at the `ChannelSpec` element to see how they should look like.
Do you mean I should save the plot as a txt file to find the ChannelSpec?
Hikmet Emre Kaya
Thank you for the valuable suggestions; it all makes perfect sense.
I had an idea about what to do when the user gives an expression for the input (By selecting objects from a JTree and putting math operations in between)
Write a separate class that reads this expression, separates it into its components
gets the common name for each object in the expression according to the string. (e.g., if it is Flux.something, get the fluxValueReference, if it is [something], get the common name for the concentration, and so on)
After converting them into their CN format, it surrounds each object with < > brackets.
It then feeds this revised formula back into the Optimization task.
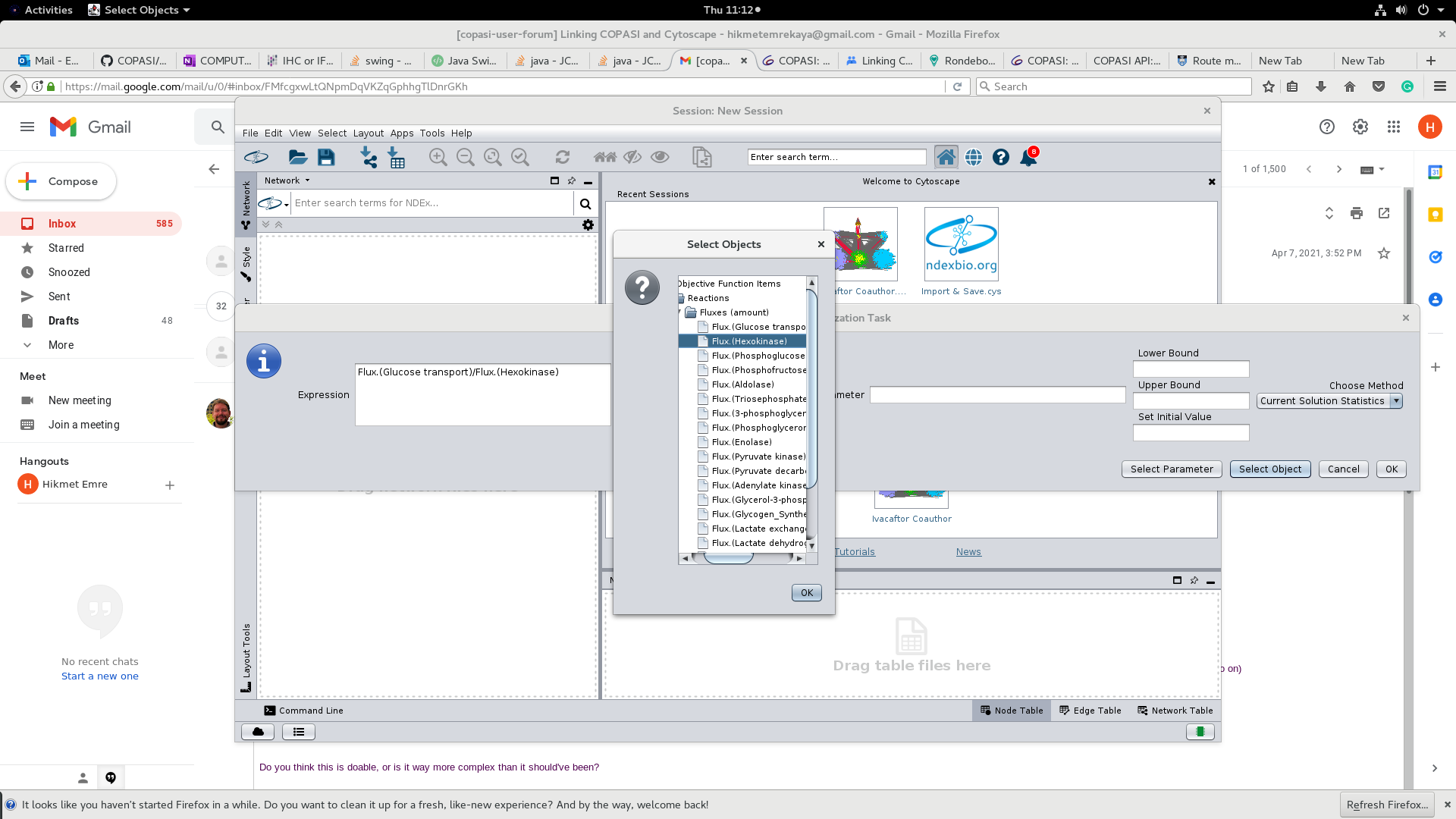
Do you think this is doable, or is it way more complex than it should've been?
Frank Bergmann
Hikmet Emre Kaya
--
You received this message because you are subscribed to a topic in the Google Groups "COPASI User Forum" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/copasi-user-forum/g_gVsfDofOE/unsubscribe.
To unsubscribe from this group and all its topics, send an email to copasi-user-fo...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/copasi-user-forum/54442d7a-3312-46d1-b812-0a3e71472e52n%40googlegroups.com.
--
Hikmet Emre Kaya
Frank Bergmann
Hikmet Emre Kaya
That definitely makes life a lot easier! To be clear, do you have to put that expression in angle brackets as well, or does COPASI recognize it when you put in optItem.setObjectiveFunction(my expression)
where my expression is: say, { (Glucose Transport).Flux } / { (Hexokinase).Flux }
Frank Bergmann
You received this message because you are subscribed to the Google Groups "COPASI User Forum" group.
To unsubscribe from this group and stop receiving emails from it, send an email to copasi-user-fo...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/copasi-user-forum/a501d7a9-9e55-4c35-8c47-13034e1b8c7dn%40googlegroups.com.
Hikmet Emre Kaya
So, let's say I got this user input with the curly brackets and passed it onto the
public void run (... ...)
- Loading the model from string
- Running time course or steady state
- Creating an optimization task
Will I have to convert that expression into the <cn1>/<cn2> format before I set the objective function in my COptProblem?
Hikmet Emre Kaya
I gave it a try and wrote the following by looking at java bindings examples:
CModelValue variableModelValue = model.createModelValue("V");
variableModelValue.setStatus(CModelEntity.Status_ASSIGNMENT);
variableModelValue.setExpression(expression);
Where, the expression could be, for example = {[ADP]}/{[ATP]}
String objectiveFunction = variableModelValue.getObject(new CCommonName("Reference=Value")).getCN().getString();
When I print out objectiveFunction, I get the following:
Objective Function: CN=Root,Model=Karlstaedt2019 - G6P accumulation via Phosphoglucose isomerase inhibition in heart muscles,Vector=Values[V],Reference=Value
But, when I enter the same expression on COPASI and look at the .cps file, the objective function is:
<ParameterText name="ObjectiveExpression" type="expression">
<CN=Root,Model=Karlstaedt2019 - G6P accumulation via Phosphoglucose isomerase inhibition in heart muscles,Vector=Compartments[cytosol],Vector=Metabolites[ADP],Reference=Concentration>/<CN=Root,Model=Karlstaedt2019 - G6P accumulation via Phosphoglucose isomerase inhibition in heart muscles,Vector=Compartments[cytosol],Vector=Metabolites[ATP],Reference=Concentration>
</ParameterText>
Does this mean, I am not getting the objective function correctly?
Hikmet Emre Kaya
Sorry for asking too many questions :D I actually managed to do the conversion from object display name to Common Name, which gives the correct value for the minimum objective function.
My question is about the optimization task itself. I am confused as to why COPASI gives a different best parameter value (corresponding to the minimum objective function) after each optimization run. Is there a document that you can suggest for me to understand the logic of optimization better?
Best,
Emre
Frank Bergmann
Hikmet Emre Kaya
To view this discussion on the web visit https://groups.google.com/d/msgid/copasi-user-forum/f12e76df-5bf1-4ae7-9afd-d47a9b1ff0e7n%40googlegroups.com.
Frank Bergmann
To view this discussion on the web visit https://groups.google.com/d/msgid/copasi-user-forum/CAFJfJ8jQNnBNqwhiNcT%2B3mO2XRCN08%3Dx-p6Rgp-YVxnxgZJtuw%40mail.gmail.com.
Pedro Mendes
There are many reasons why the best value of a run may be different from
another run. First you should check if "randomize start values" is
checked. If it is, then each run starts from a different place and
therefore algorithms are very likely to end up in different places
(including the deterministic algorithms).
If the initial values are _not_ randomized then you should only expect
that deterministic algorithms should end in the same place. These
algorithms are Levenberg-Marquardt, Truncated Newton, steepest descent,
NL2SOL, Praxis, Hooke-Jeeves and Nelder-Mead.
However the global optimization algorithms are based on stochasticity
and do not necessarily end up in the same value. Methods in this class
are all teh evolutionary algorithms (including genetic algorithm,
differential evolution, SRES, etc.), particle swarm and simulated
annealing.
A long time ago I published a paper on this topic (which was still for
my older Gepasi software) which talks a little about the algorithms.
Mendes P, Kell DB (1998) Non-linear optimization of biochemical
pathways: applications to metabolic engineering and parameter
estimation. Bioinformatics 14:869–883
https://doi.org/10.1093/bioinformatics/14.10.869
Pedro
On 11/4/21 4:17 AM, Hikmet Emre Kaya wrote:
> Hi Frank,
>
> Sorry for asking too many questions :D I actually managed to do the
> conversion from object display name to Common Name, which gives the
> correct value for the minimum objective function.
>
> My question is about the optimization task itself. I am confused as to
> why COPASI gives a different best parameter value (corresponding to the
> minimum objective function) after each optimization run. Is there a
> document that you can suggest for me to understand the logic of
> optimization better?
>
> Best,
>
> Emre
>
> On Friday, October 29, 2021 at 12:55:23 PM UTC+2 Hikmet Emre Kaya wrote:
>
> Hello again, Frank;
>
> I gave it a try and wrote the following by looking at java bindings
> examples:
>
> CModelValue variableModelValue = model.createModelValue("V");
> variableModelValue.setStatus(CModelEntity.Status_ASSIGNMENT);
>
> variableModelValue.setExpression(expression);
>
> Where, the expression could be, for example = {[ADP]}/{[ATP]}
>
> String objectiveFunction = variableModelValue.getObject(new
> CCommonName("Reference=Value")).getCN().getString();
>
> When I print out objectiveFunction, I get the following:
>
> muscles,Vector=Values[V],Reference=Value
> file, the objective function is:
>
> Phosphoglucose isomerase inhibition in heart
> muscles,Vector=Compartments[cytosol],Vector=Metabolites[ADP],Reference=Concentration>/<CN=Root,Model=Karlstaedt2019
> - G6P accumulation via Phosphoglucose isomerase inhibition in heart
> muscles,Vector=Compartments[cytosol],Vector=Metabolites[ATP],Reference=Concentration>
> Write a separate class that
> reads this expression, separates
> it into its components
>
> gets the common name for each
> object in the expression
> according to the string. (e.g.,
> if it is Flux.something, get the
> fluxValueReference, if it is
> [something], get the common name
> for the concentration, and so on)
>
> After converting them into their
> CN format, it surrounds each
> object with < > brackets.
>
>
>
>
> --
> *Hikmet Emre Kaya*
> *
> *
> *Professional Science & Wellness Writer*
> *
> *
> University of Cape Town
>
>
>
>
>
> --
> *Hikmet Emre Kaya*
> *
> *
> *Professional Science & Wellness Writer*
> *
> *
> University of Cape Town
>
>
> --
>
> You received this message because you are subscribed to
> the Google Groups "COPASI User Forum" group.
>
> To unsubscribe from this group and stop receiving emails
> from it, send an email to
> copasi-user-fo...@googlegroups.com.
> To view this discussion on the web visit
> https://groups.google.com/d/msgid/copasi-user-forum/a501d7a9-9e55-4c35-8c47-13034e1b8c7dn%40googlegroups.com
> --
> You received this message because you are subscribed to the Google
> Groups "COPASI User Forum" group.
> To unsubscribe from this group and stop receiving emails from it, send
> an email to copasi-user-fo...@googlegroups.com
> <https://groups.google.com/d/msgid/copasi-user-forum/e9a6b2ab-faa5-4709-9907-d402434d7604n%40googlegroups.com?utm_medium=email&utm_source=footer>.
Hikmet Emre Kaya
Thank you for your insightful comments. These suggestions will definitely help me grasp the logic of optimization much better.
I have one more question related to optimization. In stand alone COPASI, the best value is shown in a live-updating plot, but I am not sure if I can generate the same plot from the Java bindings. It seems that one can only retrieve the final value after the task is finished. Is there a way to iteratively access the value found after each function evaluation?
Frank Bergmann
I have one more question related to optimization. In stand alone COPASI, the best value is shown in a live-updating plot, but I am not sure if I can generate the same plot from the Java bindings. It seems that one can only retrieve the final value after the task is finished. Is there a way to iteratively access the value found after each function evaluation?
Hikmet Emre Kaya
I have also been working on bringing the Parameter Scan feature into Cytoscape. Currently working on single parameter scan for now. The task does run without errors, but I am confused about retrieving the set of results for each parameter. For example, the user makes a selection from a plot assistant to get steady state concentration for each parameter value and create a plot (as described here: https://www.youtube.com/watch?v=mRPY3m8pWJU).
Do you need to implement the callback class idea here as well?
Frank Bergmann
Hikmet Emre Kaya
Thank you for the detailed response. I actually have been trying to create a report file, but it's coming out empty for some reason. Let me go over this example, and I'll get back to you if I have any questions.
Best,
Emre
Hikmet Emre Kaya
There is a part that confuses me about this example. Under create_datahandler, there is the line:
if isinstance(obj, COPASI.CModel): # fix for time
obj = obj.getValueReference()
in java, obj = dm.findObjectByDisplayName(name) is a CDataObject, and it does not have the getValueReference option. So, I am not sure how this part translates to Java.
Hikmet Emre Kaya
Frank Bergmann
--
You received this message because you are subscribed to a topic in the Google Groups "COPASI User Forum" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/copasi-user-forum/g_gVsfDofOE/unsubscribe.
To unsubscribe from this group and all its topics, send an email to copasi-user-fo...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/copasi-user-forum/bc0e9a69-25d3-46bc-b073-808abcd12d4cn%40googlegroups.com.
Frank Bergmann
--
You received this message because you are subscribed to a topic in the Google Groups "COPASI User Forum" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/copasi-user-forum/g_gVsfDofOE/unsubscribe.
To unsubscribe from this group and all its topics, send an email to copasi-user-fo...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/copasi-user-forum/01206f32-cb01-4afd-84e4-2fcb24b79184n%40googlegroups.com.
Hikmet Emre Kaya
I am using this model as an example.
In the parameter scan, I chose the "scan" option, specify one of the reaction parameters as my parameter object (e.g., (Phosphofructosekinase).Vmax_4 with intervals=10; min = 40, and max=160). Then, I click on plot assistant and select Scan of Concentrations, Volumes, and Global Quantity Values. Then, I go to the plot itself and select one of the species (e.g., [ATP]) and delete all the other curves. For the subtask, I chose steady state so that I can monitor how the std st concentration of that metabolite changes throughout the parameter interval. As far as I understand, COPASI then repeats the subtask for each interval by incrementing the parameter value between the min and the max. Then, you generate a plot like the one attached.
So, I am wanting to collect that specific model variable for each parameter value to see its dependency on that parameter.
Hikmet Emre Kaya
parameterGroup.addParameter("Number of Intervals", CCopasiParameter.Type_UINT);
CCopasiParameter parameter = parameterGroup.getParameter("Number of Intervals");
parameter.setUIntValue(some value);
Then, I added the parameter to be scanned:
parameterGroup.addParameter("Object", CCopasiParameter.CN);
parameter=parameterGroup.getParameter("Object")
parameter.setStringValue(the string representation of the CN of that object);
I also defined the subtask in a similar way
Frank Bergmann
1.301643024627298 0.642530054163137
1.6932150133907584 0.5535842632054738
1.879352731470603 0.48802610343669633
1.9900205515407214 0.4414058753996679
2.0657261238718307 0.40597339993295095
2.122188999735226 0.3775488429123931
2.166767986701773 0.3538312767697282
2.2033979691565624 0.3334578978875124
2.2343940840051046 0.315566299836136
2.2612189291772085 0.2995804283474704
2.284848426245759 0.2850983600572651
2.284848426245759 0.2850983600572651
You received this message because you are subscribed to the Google Groups "COPASI User Forum" group.
To unsubscribe from this group and stop receiving emails from it, send an email to copasi-user-fo...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/copasi-user-forum/64bd8ef2-5abf-4f51-b5bc-6491e9bafe05n%40googlegroups.com.
Hikmet Emre Kaya
Thanks! This clarifies it a lot. Currently, I am getting the error "Internal problem with scan definition". from the statement
if (!scanTask.initializeRawWithOutputHandler((int)CCopasiTask.OUTPUT_UI, dh))
It makes me think I am making a mistake when specifying the parameters. I was using the code snippet from the Java Bindings.txt as an example
CScanTask scanTask=
(CScanTask)CDataModel.getGlobal().addTask(CCopasiTask.scan);
CScanProblem scanProblem=(CScanProblem)scanTask.getProblem();
// set the desired subtask
scanProblem.setSubtask(CCopasiTask.timeCourse);
// always use the same initial values
scanProblem.setAdjustInitialConditions(false);
// get the ScanItems parameter group from the problem
CCopasiParameterGroup scanItems=scanProblem.getGroup("ScanItems");
// create a new parameter group to hold a scan item
CCopasiParameterGroup scanItem=new CCopasiParameterGroup("scanItem");
// add the parameters "Number of steps", "Type" and "Object" to the
// scan item and set their values
parameterGroup.addParameter("Number of steps",
CCopasiParameter.UINT);
CCopasiParameter parameter=parameterGroup.getParameter("Number of steps");
parameter..setUIntValue(10);
parameterGroup.addParameter("Type", CCopasiParameter.UINT);
parameter=parameterGroup.getParameter("Type")
parameter.setUIntValue(CScanProblem.SCAN_REPEAT);
parameterGroup.addParameter("Object", CCopasiParameter.CN);
parameter=parameterGroup.getParameter("Object") parameter.setStringValue("");
here, I am confused as to where the parameters are being added. Where was the parameterGroup instance created? Why are scanItems and scanItem not used for parameter addition in the code above?
Frank Bergmann
To view this discussion on the web visit https://groups.google.com/d/msgid/copasi-user-forum/55464dfe-197a-466c-8cec-b1288e46d493n%40googlegroups.com.
Hikmet Emre Kaya
To view this discussion on the web visit https://groups.google.com/d/msgid/copasi-user-forum/CAEnSYuJidpjLAt%3DZ1Tuy4RY9UcQ5ecfQWXjZttk8Sf7adrHcgw%40mail.gmail.com.
--
Frank Bergmann
Hikmet Emre Kaya
I hope this message finds you well.
I have been making a lot of progress with linking COPASI and Cytoscape to utilize Cytoscape's visualization and database options.
I have a quick question. How does one update an initial value (e.g., initial concentration) of an already existing model entity.
I get the concept of creating a vector to store all the changes.
For example, if I want to change the initial concentration of a metabolite, I would do the following:
ObjectStdVector changedObjects = new ObjectStdVector();
model.getMetabolite(i).setInitialConcentration(...
changedObjects.add(model.getMetabolite(i).getInitialConcentrationReference());
model.CompileIfNecessary();
model.updateInitialValues(changedObjects);
However, the setInitialConcentration method does not seem to change the initial concentration. How does one go about this? I would appreciate it if you could kindly advise.
Regards;
Emre
Hikmet Emre Kaya
I actually figured it out. I needed to use the compileIsInitialValueChangeAllowedMethod() of CMetab.
Frank Bergmann
--
You received this message because you are subscribed to the Google Groups "COPASI User Forum" group.
To unsubscribe from this group and stop receiving emails from it, send an email to copasi-user-fo...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/copasi-user-forum/d607ce10-47a6-4382-8977-53a0ca3b1e5dn%40googlegroups.com.
Hikmet Emre Kaya
The code does work for changing .cps files. However, I am running into some issues when I import an SBML file, make changes and save it (and export it).
After making changes in an sbml file and exporting the changed file as SBML, when I need to import it again, I receive the following error.
java.lang.RuntimeException: C++ CCopasiException exception thrown
at org.COPASI.COPASIJNI.CDataModel_importSBML__SWIG_2(Native Method)
...
How does one go about importing an SBML file, changing its values, and saving the new model as an SBML file?
Thanks for your assistance,
Emre
Frank Bergmann
Hikmet Emre Kaya
Hi Frank,
Below is the CCopasiMessage.getAllMessageText()
All this error shows up when I modify an initial concentration value and re-export the SBML file. The next time I import it, I receive the error .
error: SBML (40): LIBSBML ERROR 10215 at line 2862 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'Km2ATP' in the math element of the <kineticLaw> uses 'Km2ATP' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 2862 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'Km2Glc' in the math element of the <kineticLaw> uses 'Km2Glc' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 2862 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'Ks2Glc' in the math element of the <kineticLaw> uses 'Ks2Glc' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 2862 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'Vm2' in the math element of the <kineticLaw> uses 'Vm2' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 2926 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'K3Gly' in the math element of the <kineticLaw> uses 'K3Gly' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 2926 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'Km30' in the math element of the <kineticLaw> uses 'Km30' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 2926 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'Km3G6P' in the math element of the <kineticLaw> uses 'Km3G6P' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 2926 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'Vm3' in the math element of the <kineticLaw> uses 'Vm3' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 2926 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'n3' in the math element of the <kineticLaw> uses 'n3' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 2996 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'K4AMP' in the math element of the <kineticLaw> uses 'K4AMP' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 2996 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'K4ATP' in the math element of the <kineticLaw> uses 'K4ATP' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 2996 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'K4F6P' in the math element of the <kineticLaw> uses 'K4F6P' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 2996 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'L40' in the math element of the <kineticLaw> uses 'L40' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 2996 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'Vm4' in the math element of the <kineticLaw> uses 'Vm4' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 2996 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'c4AMP' in the math element of the <kineticLaw> uses 'c4AMP' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 2996 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'c4ATP' in the math element of the <kineticLaw> uses 'c4ATP' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 2996 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'c4F6P' in the math element of the <kineticLaw> uses 'c4F6P' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 2996 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'g4R' in the math element of the <kineticLaw> uses 'g4R' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 2996 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'gT' in the math element of the <kineticLaw> uses 'gT' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3120 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'K5ADP' in the math element of the <kineticLaw> uses 'K5ADP' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3120 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'K5AMP' in the math element of the <kineticLaw> uses 'K5AMP' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3120 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'K5ATP' in the math element of the <kineticLaw> uses 'K5ATP' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3120 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'K5G3P' in the math element of the <kineticLaw> uses 'K5G3P' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3120 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'K5NAD' in the math element of the <kineticLaw> uses 'K5NAD' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3120 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'K5NADH' in the math element of the <kineticLaw> uses 'K5NADH' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3120 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'NAD' in the math element of the <kineticLaw> uses 'NAD' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3120 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'NADH' in the math element of the <kineticLaw> uses 'NADH' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3120 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'Vm5' in the math element of the <kineticLaw> uses 'Vm5' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3186 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'K6ADP' in the math element of the <kineticLaw> uses 'K6ADP' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3186 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'K6FDP' in the math element of the <kineticLaw> uses 'K6FDP' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3186 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'K6PEP' in the math element of the <kineticLaw> uses 'K6PEP' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3186 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'L60' in the math element of the <kineticLaw> uses 'L60' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3186 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'Vm6' in the math element of the <kineticLaw> uses 'Vm6' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3186 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'c6ADP' in the math element of the <kineticLaw> uses 'c6ADP' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3186 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'c6FDP' in the math element of the <kineticLaw> uses 'c6FDP' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3186 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'c6PEP' in the math element of the <kineticLaw> uses 'c6PEP' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3186 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'g6R' in the math element of the <kineticLaw> uses 'g6R' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3186 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'g6T' in the math element of the <kineticLaw> uses 'g6T' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3186 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'h6' in the math element of the <kineticLaw> uses 'h6' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3186 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'q6' in the math element of the <kineticLaw> uses 'q6' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3246 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'K6ADP' in the math element of the <kineticLaw> uses 'K6ADP' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3246 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'K6FDP' in the math element of the <kineticLaw> uses 'K6FDP' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3246 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'K6PEP' in the math element of the <kineticLaw> uses 'K6PEP' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3246 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'L60' in the math element of the <kineticLaw> uses 'L60' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3246 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'Vm7' in the math element of the <kineticLaw> uses 'Vm7' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3246 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'c6ADP' in the math element of the <kineticLaw> uses 'c6ADP' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3246 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'c6FDP' in the math element of the <kineticLaw> uses 'c6FDP' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3246 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'c6PEP' in the math element of the <kineticLaw> uses 'c6PEP' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3246 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'g6R' in the math element of the <kineticLaw> uses 'g6R' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3246 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'g6T' in the math element of the <kineticLaw> uses 'g6T' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3246 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'h6' in the math element of the <kineticLaw> uses 'h6' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3246 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'q6' in the math element of the <kineticLaw> uses 'q6' that is not the id of a species/compartment/parameter/reaction.
.
SBML (40): LIBSBML ERROR 10215 at line 3305 column 8: Outside of a <functionDefinition>, if a <ci> element is not the first element within a MathML <apply>, then the <ci>'s value can only be chosen from the set of identifiers of (in L2V1) <species>, <compartment>, or <parameter> objects; (in L2V2-L2V5), <species>, <compartment>, <parameter> or <reaction> objects; (in L3V1) <species>, <compartment>, <parameter>, <reaction> or <speciesReference> objects and (in L3V2) <species>, <compartment>, <parameter>, <reaction>, <speciesReference> objects or L3 package objects with defined mathematical meaning.
Reference: L2V4 Section 3.4.3
The formula 'Vm8' in the math element of the <kineticLaw> uses 'Vm8' that is not the id of a species/compartment/parameter/reaction.
.
>EXCEPTION 2022-02-14T15:24:06<
SBML (27): Error in kinetic law for reaction 'Hexokinase'.
>EXCEPTION 2022-02-14T15:24:06<
Error while importing reaction "Vhk".
SBML (27): Error in kinetic law for reaction 'Hexokinase'.
Frank Bergmann
Hikmet Emre Kaya
Error while loading file /home/people/hkaya/Downloads/CardioGlyco-v1.xml!
>EXCEPTION 2022-02-14T16:05:04<
Error while importing reaction "HXT".
SBML (27): Error in kinetic law for reaction 'Glucose transport'.
So, it's actually not the code itself causing the error, I think. It has to do with how one imports, modifies and saves and SBML in general. I tried this with other sbml files (.xml) and even tried importing and exporting without changing anything. It still breaks the file somehow.
Let me know what you think.
To view this discussion on the web visit https://groups.google.com/d/msgid/copasi-user-forum/16a2d992-f1c2-4035-b9f4-efe860f32558n%40googlegroups.com.
Frank Bergmann
To view this discussion on the web visit https://groups.google.com/d/msgid/copasi-user-forum/CAFJfJ8giRXyLABkw%3Do%2BVqqgUORCb-sSinfV53UMSaiQM0tTcpQ%40mail.gmail.com.
Hikmet Emre Kaya
openjdk 11.0.9 2020-10-20 LTS
OpenJDK Runtime Environment 18.9 (build 11.0.9+11-LTS)
OpenJDK 64-Bit Server VM 18.9 (build 11.0.9+11-LTS, mixed mode, sharing) (I am not sure if this is what you are asking)
Also, the alternative method is causing Cytoscape to crash with the following error:
# A fatal error has been detected by the Java Runtime Environment:
#
# SIGSEGV (0xb) at pc=0x0000000000000000, pid=11511, tid=11535
#
# JRE version: OpenJDK Runtime Environment 18.9 (11.0.9+11) (build 11.0.9+11-LTS)
# Java VM: OpenJDK 64-Bit Server VM 18.9 (11.0.9+11-LTS, mixed mode, tiered, compressed oops, g1 gc, linux-amd64)
# Problematic frame:
# C 0x0000000000000000
#
# No core dump will be written. Core dumps have been disabled. To enable core dumping, try "ulimit -c unlimited" before starting Java again
#
# An error report file with more information is saved as:
# /home/people/hkaya/Cytoscape_v3.8.2/framework/hs_err_pid11511.log
#
# If you would like to submit a bug report, please visit:
# https://bugzilla.redhat.com/enter_bug.cgi?product=Red%20Hat%20Enterprise%20Linux%207&component=java-11-openjdk
# The crash happened outside the Java Virtual Machine in native code.
# See problematic frame for where to report the bug.
#
./framework/bin/karaf: line 177: 11511 Aborted ${KARAF_EXEC} "${JAVA}" ${JAVA_OPTS} --add-reads=java.xml=java.logging --add-exports=java.base/org.apache.karaf.specs.locator=java.xml,ALL-UNNAMED --patch-module java.base=$KARAF_HOME/lib/endorsed/org.apache.karaf.specs.locator-4.2.4.jar --patch-module java.xml=$KARAF_HOME/lib/endorsed/org.apache.karaf.specs.java.xml-4.2.4.jar -p $KARAF_HOME/lib/openjfx/$JAVAFX_DIR --add-modules javafx.base,javafx.swing,javafx.media,javafx.web --add-opens java.base/java.security=ALL-UNNAMED --add-opens java.base/java.net=ALL-UNNAMED --add-opens java.base/java.lang=ALL-UNNAMED --add-opens java.base/java.util=ALL-UNNAMED --add-opens java.naming/javax.naming.spi=ALL-UNNAMED --add-opens java.rmi/sun.rmi.transport.tcp=ALL-UNNAMED --add-exports=java.base/sun.net.www.protocol.http=ALL-UNNAMED --add-exports=java.base/sun.net.www.protocol.https=ALL-UNNAMED --add-exports=java.base/sun.net.www.protocol.jar=ALL-UNNAMED --add-exports=jdk.xml.dom/org.w3c.dom.html=ALL-UNNAMED --add-exports=jdk.naming.rmi/com.sun.jndi.url.rmi=ALL-UNNAMED -Dkaraf.instances="${KARAF_HOME}/instances" -Dkaraf.home="${KARAF_HOME}" -Dkaraf.base="${KARAF_BASE}" -Dkaraf.data="${KARAF_DATA}" -Dkaraf.etc="${KARAF_ETC}" -Dkaraf.log="${KARAF_LOG}" -Dkaraf.restart.jvm.supported=true -Djava.io.tmpdir="${KARAF_DATA}/tmp" -Djava.util.logging.config.file="${KARAF_BASE}/etc/java.util.logging.properties" ${KARAF_SYSTEM_OPTS} ${KARAF_OPTS} ${OPTS} -classpath "${CLASSPATH}" ${MAIN} "$@"
Frank Bergmann
You received this message because you are subscribed to a topic in the Google Groups "COPASI User Forum" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/copasi-user-forum/g_gVsfDofOE/unsubscribe.
To unsubscribe from this group and all its topics, send an email to copasi-user-fo...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/copasi-user-forum/9eb79323-202f-47e0-adb6-0ba2761d7e7dn%40googlegroups.com.
Hikmet Emre Kaya
I have downloaded and installed the new Java Bindings, thanks!
When the sbml file is of the format L2V1, then the exportSBML function works perfectly in both COPASI and its java bindings.
Example: https://www.ebi.ac.uk/biomodels/BIOMD0000000061
when the format is L2V4: export SBML still works perfectly in standalone COPASI, I can change a model value, and the change will be saved in the exported file.
With the java bindings version, the exportSBML method will not register any changes made in the model. I definitely made sure overwrite is possible by writing dm.exportModel(modelName, true)
Example: https://www.ebi.ac.uk/biomodels/BIOMD0000000063
Is there a specific issue in the exportSBML java bindings version that allows modifications only in certain SBML versions?
Perhaps you could try to import, change model values (e.g., some initial concentration), and export the above .xml files in java bindings and let me know if you run into the same issue?
Hikmet Emre Kaya
Frank Bergmann
Frank Bergmann
Hikmet Emre Kaya
Also, what OS are you using? I am having trouble opening the app in Mac because it crashes when I try to import files. I copied the .so file to java.library.path, but still did not manage to solve this. Do you have any idea? I have not had this issue in Linus CentOS or Windows.
Frank Bergmann
Hikmet Emre Kaya
Hikmet Emre Kaya
Hikmet Emre Kaya
model.getMetabolite(i).setInitialConcentration(...
changedObjects.add(model.getMetabolite(i).getInitialConcentrationReference());
model.updateInitialValues(changedObjects);
model.compileIfNecessary();
model.getMetabolite(i).setStatus(...);
model.updateInitialValues(changedObjects);
model.compileIfNecessary();
Hikmet Emre Kaya
Frank Bergmann
Hikmet Emre Kaya
for (int j = 0; j < data.size(); j++)
{
System.out.print(data.get(j));
if (j + 1 < data.size())
System.out.print("\t");
}
}
To view this discussion on the web visit https://groups.google.com/d/msgid/copasi-user-forum/d0dc3611-c932-46b7-95c7-f92ad20620e6n%40googlegroups.com.
Frank Bergmann
Hikmet Emre Kaya
--
You received this message because you are subscribed to a topic in the Google Groups "COPASI User Forum" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/copasi-user-forum/g_gVsfDofOE/unsubscribe.
To unsubscribe from this group and all its topics, send an email to copasi-user-fo...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/copasi-user-forum/f6db4993-9b13-4c75-b9f6-8113191814ecn%40googlegroups.com.
Frank Bergmann
Hikmet Emre Kaya
--
You received this message because you are subscribed to a topic in the Google Groups "COPASI User Forum" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/copasi-user-forum/g_gVsfDofOE/unsubscribe.
To unsubscribe from this group and all its topics, send an email to copasi-user-fo...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/copasi-user-forum/ce961880-8bfb-4b11-8200-9d7e2516b9bdn%40googlegroups.com.
Hikmet Emre Kaya
Frank Bergmann
Hikmet Emre Kaya
--
You received this message because you are subscribed to a topic in the Google Groups "COPASI User Forum" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/copasi-user-forum/g_gVsfDofOE/unsubscribe.
To unsubscribe from this group and all its topics, send an email to copasi-user-fo...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/copasi-user-forum/22064fda-0759-49e6-9889-c0beecaebe66n%40googlegroups.com.
Frank Bergmann
You received this message because you are subscribed to the Google Groups "COPASI User Forum" group.
To unsubscribe from this group and stop receiving emails from it, send an email to copasi-user-fo...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/copasi-user-forum/CAFJfJ8geqBanFd864EOaphcFwTUCCG0csnNyJ2yUs03bpCkdyA%40mail.gmail.com.
