Skip to first unread message
Hugo Harada
Jul 18, 2018, 12:15:52 AM7/18/18
to lavaan
Hello everyone,
I am trying to show the equivalence of the IRT parameter estimation done in MIRT and in LAVAAN. I was successful in the unidimensional case as the results below demonstrate. (see code below and attached)
a_sem a_triItem_1 0.7188273 0.8250552Item_2 0.7355265 0.7230608Item_3 0.9084112 0.8899989Item_4 0.6917957 0.6886588Item_5 0.6194848 0.6575904b_sem b_triItem_1|t1 -3.6782281 -3.3607460Item_2|t1 -1.3862522 -1.3695513Item_3|t1 -0.2827214 -0.2798928Item_4|t1 -1.8995476 -1.8657302Item_5|t1 -3.2898952 -3.1229745
But I am having problems defining a runnable LAVAAN model for the bidimensional case. I expect per item: one threshold value and two lambda (factor) values (one per dimension). Is the syntax below wrong?
lavaan.lsat6.2pl.model.2D <-'Theta1 =~ l11*Item_1 + l12*Item_2 + l13*Item_3 + l14*Item_4 + l15*Item_5Theta2 =~ l21*Item_1 + l22*Item_2 + l23*Item_3 + l24*Item_4 + l25*Item_5Item_1 | th1 *t1Item_2 | th2 *t1Item_3 | th3 *t1Item_4 | th4 *t1Item_5 | th5 *t1'
Below I try to reproduce the results shown in by McDonald in Normal Ogive Multidimensional Model (1985). Can anyone help?
Any insights will be highly appreciated.
Hugo
######################################################################################
######################################################################################
rm(list=ls())
library(lavaan)
library(mirt)
dat <- expand.table(LSAT6)
head(dat)
#descript(LSAT6)
##########################################################################
# Unidimensional case - No problem here.
##########################################################################
#estimating parameters using MIRT.
mirt.lSAT6.2PL = mirt(dat, 1, itemtype = '2PL', method = 'EM') #
PAR=coef(mirt.lSAT6.2PL,simplify=TRUE,IRTpars=TRUE)
PAR$items[,1]/1.7 #normal ogive scaling
PAR
#estimating parameters using lavaan.
lavaan.lsat6.2pl.model <-'
Theta =~ l1*Item_1 + l2*Item_2 + l3*Item_3 + l4*Item_4 + l5*Item_5
Item_1 | th1 *t1
Item_2 | th2 *t1
Item_3 | th3 *t1
Item_4 | th4 *t1
Item_5 | th5 *t1
'
#fitting model
lavaan.lsat6.2pl.model.fit <- cfa(lavaan.lsat6.2pl.model, data = dat , std.lv=TRUE , ordered =c("Item_1","Item_2","Item_3","Item_4","Item_5"))
#getting lambda values
lambda <- lavInspect(lavaan.lsat6.2pl.model.fit,what = 'est')$lambda
#getting tau values
tau <- lavInspect(lavaan.lsat6.2pl.model.fit,what = 'est')$tau
# Convert regression to IRT
# alpha1 := ( l1)/ sqrt (1- l1^2)
# beta1 := ( - th1 )/ sqrt (1- l1^2)
# a_IRT := alpha1*1.7
# b_IRT := -beta1/alpha1
a_sem <- (lambda/sqrt(1-lambda*lambda))
b_sem <- tau/sqrt(1-lambda*lambda)/a_sem
a_sem <- 1.7*a_sem #normal ogive scaling
#making comparison tables. a and b values are close.
a_comp <- cbind(a_sem,PAR$items[,1])
colnames(a_comp)<-c("a_sem","a_tri")
a_comp
b_comp <- cbind(b_sem,PAR$items[,2])
colnames(b_comp)<-c("b_sem","b_tri")
b_comp
##########################################################################
# Bidimensional case -
##########################################################################
#estimating parameters using MIRT.
mirt.lSAT6.2PL.2D = mirt(dat, 2, itemtype = '2PL', method = 'EM') #
PAR=coef(mirt.lSAT6.2PL.2D,simplify=TRUE)
PAR
#MIRT results
# a1 a2 d g u
# Item_1 0.632 0.783 2.898 0 1
# Item_2 0.574 0.465 0.994 0 1
# Item_3 0.749 0.510 0.250 0 1
# Item_4 1.228 -0.402 1.526 0 1
# Item_5 0.685 0.000 2.066 0 1
#estimating bidimensional parameters using lavaan. ??
#estimating parameters using lavaan.
lavaan.lsat6.2pl.model.2D <-'
Theta1 =~ l11*Item_1 + l12*Item_2 + l13*Item_3 + l14*Item_4 + l15*Item_5
Theta2 =~ l21*Item_1 + l22*Item_2 + l23*Item_3 + l24*Item_4 + l25*Item_5
Item_1 | th1 *t1
Item_2 | th2 *t1
Item_3 | th3 *t1
Item_4 | th4 *t1
Item_5 | th5 *t1
'
lavaan.lsat6.2pl.model.2D.fit <- cfa(lavaan.lsat6.2pl.model.2D, data = dat , std.lv=TRUE , ordered =c("Item_1","Item_2","Item_3","Item_4","Item_5"))
## Models is not identifiable?
Terrence Jorgensen
Jul 18, 2018, 8:56:37 AM7/18/18
to lavaan
## Models is not identifiable?
I think it is not identified because by default, cfa() will freely estimate the factor correlation because CFAs usually have simple structure or at least some loadings fixed to zero, which overcomes the rotational indeterminacy issue in EFAs. To freely estimate all loadings/discriminations, the factor correlation(s) must be fixed (typically to zero) to identify the model. You can specify this in the syntax with Theta1 ~~ 0*Theta2, or by using setting argument orthogonal=TRUE (see the ?lavOptions help page).
Terrence D. Jorgensen
Postdoctoral Researcher, Methods and Statistics
Research Institute for Child Development and Education, the University of Amsterdam
UvA web page: http://www.uva.nl/profile/t.d.jorgensen
Hugo Harada
Jul 18, 2018, 12:11:56 PM7/18/18
to lav...@googlegroups.com
Hi Terence,
Thank you for your comments. I tried your suggestions and I got the following warning
Warning message:In lav_model_vcov(lavmodel = lavmodel, lavsamplestats = lavsamplestats, :lavaan WARNING: could not compute standard errors!lavaan NOTE: this may be a symptom that the model is not identified.
and I also notice that lavaan was not able to differentiate between the two dimensions at all. The factor loadings are exactly the same for theta1 and theta2.
Theta1 =~Item_1 (l11) 0.275 NA 0.275 0.275Item_2 (l12) 0.281 NA 0.281 0.281Item_3 (l13) 0.333 NA 0.333 0.333Item_4 (l14) 0.267 NA 0.267 0.267Item_5 (l15) 0.242 NA 0.242 0.242Theta2 =~Item_1 (l21) 0.275 NA 0.275 0.275Item_2 (l22) 0.281 NA 0.281 0.281Item_3 (l23) 0.333 NA 0.333 0.333Item_4 (l24) 0.267 NA 0.267 0.267Item_5 (l25) 0.242 NA 0.242 0.242
lavaan.lsat6.2pl.model.2D <-'Theta1 =~ l11*Item_1 + l12*Item_2 + l13*Item_3 + l14*Item_4 + l15*Item_5Theta2 =~ l21*Item_1 + l22*Item_2 + l23*Item_3 + l24*Item_4 + l25*Item_5Item_1 | th1 *t1Item_2 | th2 *t1Item_3 | th3 *t1Item_4 | th4 *t1Item_5 | th5 *t1
Theta1 ~~ 0*Theta2
'lavaan.lsat6.2pl.model.2D.fit <- cfa(lavaan.lsat6.2pl.model.2D, data = dat , std.lv=TRUE , ordered =c("Item_1","Item_2","Item_3","Item_4","Item_5"))
summary ( lavaan.lsat6.2pl.model.2D.fit , standardized = TRUE )
The complete output of summary is as below. Any more ideas?
lavaan (0.5-23.1097) converged normally after 25 iterationsNumber of observations 1000Estimator DWLS RobustMinimum Function Test Statistic 4.051 4.051Degrees of freedom 0 0Minimum Function Value 0.0020255466035Scaling correction factor NAShift parameterfor simple second-order correction (Mplus variant)Parameter Estimates:Information ExpectedStandard Errors Robust.semLatent Variables:Estimate Std.Err z-value P(>|z|) Std.lv Std.allTheta1 =~Item_1 (l11) 0.275 NA 0.275 0.275Item_2 (l12) 0.281 NA 0.281 0.281Item_3 (l13) 0.333 NA 0.333 0.333Item_4 (l14) 0.267 NA 0.267 0.267Item_5 (l15) 0.242 NA 0.242 0.242Theta2 =~Item_1 (l21) 0.275 NA 0.275 0.275Item_2 (l22) 0.281 NA 0.281 0.281Item_3 (l23) 0.333 NA 0.333 0.333Item_4 (l24) 0.267 NA 0.267 0.267Item_5 (l25) 0.242 NA 0.242 0.242Covariances:Estimate Std.Err z-value P(>|z|) Std.lv Std.allTheta1 ~~Theta2 0.000 0.000 0.000Intercepts:Estimate Std.Err z-value P(>|z|) Std.lv Std.all.Item_1 0.000 0.000 0.000.Item_2 0.000 0.000 0.000.Item_3 0.000 0.000 0.000.Item_4 0.000 0.000 0.000.Item_5 0.000 0.000 0.000Theta1 0.000 0.000 0.000Theta2 0.000 0.000 0.000Thresholds:Estimate Std.Err z-value P(>|z|) Std.lv Std.allItm_1|t1 (th1) -1.433 NA -1.433 -1.433Itm_2|t1 (th2) -0.550 NA -0.550 -0.550Itm_3|t1 (th3) -0.133 NA -0.133 -0.133Itm_4|t1 (th4) -0.716 NA -0.716 -0.716Itm_5|t1 (th5) -1.126 NA -1.126 -1.126Variances:Estimate Std.Err z-value P(>|z|) Std.lv Std.all.Item_1 0.848 0.848 0.848.Item_2 0.842 0.842 0.842.Item_3 0.778 0.778 0.778.Item_4 0.858 0.858 0.858.Item_5 0.883 0.883 0.883Theta1 1.000 1.000 1.000Theta2 1.000 1.000 1.000Scales y*:Estimate Std.Err z-value P(>|z|) Std.lv Std.allItem_1 1.000 1.000 1.000Item_2 1.000 1.000 1.000Item_3 1.000 1.000 1.000Item_4 1.000 1.000 1.000Item_5 1.000 1.000 1.000
Hugo
--
You received this message because you are subscribed to the Google Groups "lavaan" group.
To unsubscribe from this group and stop receiving emails from it, send an email to lavaan+unsubscribe@googlegroups.com.
To post to this group, send email to lav...@googlegroups.com.
Visit this group at https://groups.google.com/group/lavaan.
For more options, visit https://groups.google.com/d/optout.
Hugo Harada Adaptativa Inteligência Educacional S.A. | |
Mauricio Garnier-Villarreal
Jul 18, 2018, 12:15:47 PM7/18/18
to lavaan
I think there is something intrinsically different on how CFA and IRT identify the models. At least I cant see how IRT is identifyng this model
For the CFA, based on the polychoric correlation the model has -1 df, so it needs to have one more paremeter fixed, as Terry said one way would be to fix the factor correlation to 0
But the IRT model, when look at the fit with the M2() function, it has 2 df, while estimating 10 as, 5 bs, and the factor correlation. I am not sure where is IRT getting those degrees of freedom from.
I tested this with both delta and theta parameterization in lavaan.
I think the full information vs sufficient statistic approach have different identification limits on examples like this
Hugo Harada
Jul 19, 2018, 12:16:48 AM7/19/18
to lav...@googlegroups.com
Maurício, Terence.
Thanks, Mauricio.
It seems to me that it all boils down to a EFA for the model below in which the relation between Theta1 and Theta2 are oblique.
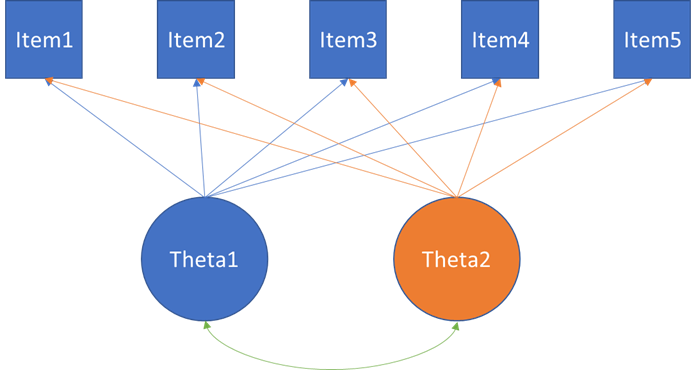
I found someone else with the same problem and he solved it but imposing anchor items with 0 loadings with no cross-loadings.. See below.
Using the same rational, I could only make the following model run. Setting arbitrarily some loadings to zero seems to be a big restriction. and I would really like to drop the orthogonal restriction. Any ideas?
lavaan.lsat6.2pl.model.2D <-'Theta1 =~ l11*Item_1 + l12*Item_2 + l13*Item_3 + 0*Item_4 + l15*Item_5Theta2 =~ l21*Item_1 + l22*Item_2 + 0*Item_3 + l24*Item_4 + l25*Item_5
Item_1 | th1 *t1Item_2 | th2 *t1Item_3 | th3 *t1Item_4 | th4 *t1Item_5 | th5 *t1
Theta1 ~~ 0*Theta2
'lavaan.lsat6.2pl.model.2D.fit <- cfa(lavaan.lsat6.2pl.model.2D,data = dat ,
std.lv=TRUE , #residual variance set to 1.0,std.ov=TRUE,ordered =c("Item_1","Item_2","Item_3","Item_4","Item_5"),debug=TRUE)
Regards,
Hugo
--
You received this message because you are subscribed to the Google Groups "lavaan" group.
To unsubscribe from this group and stop receiving emails from it, send an email to lavaan+unsubscribe@googlegroups.com.
To post to this group, send email to lav...@googlegroups.com.
Visit this group at https://groups.google.com/group/lavaan.
For more options, visit https://groups.google.com/d/optout.
Mauricio Garnier-Villarreal
Jul 19, 2018, 1:28:30 AM7/19/18
to lavaan
Hugo
The semTools package has an efa function to do EFA from the lavaan structure. So you can run the EFA with WLSMV for ordered items, with the respective rotation functions
I played with a toy example, simulated binary data for 2 factors (5 items each)
When I run the EFA for only the 5 items for factor 1 asking for 2 factors, it gives me an error of negative variance in some cases (got the same error with the LSAT data).
When I do the EFA for the 10 items which properly represents 2 factors, I get results without errors
So, I did a quick simulation, with 100 reps, and when you use 5 items that actually come from 1 factor and ask for 2 factors in the EFA, you get a negative variance 72.6% of the time
Here is the code to do the EFA with semTools with WLS. The efaUnrotate function can take other lavaan arguments
### run EFA with WLSMV for ordered items
ef2_irt <- efaUnrotate(data=dat,estimator="wlsmv",nf=2,
ordered =colnames(dat))
summary(ef2_irt, std = TRUE)
inspect(ef2_irt, "std")
## use oblique rotation
ef2_ob <- oblqRotate(ef2_irt)
summary(ef2_ob,suppress=.001)
To unsubscribe from this group and stop receiving emails from it, send an email to lavaan+un...@googlegroups.com.
To post to this group, send email to lav...@googlegroups.com.
Visit this group at https://groups.google.com/group/lavaan.
For more options, visit https://groups.google.com/d/optout.
Hugo Harada
Jul 19, 2018, 10:54:10 AM7/19/18
to lav...@googlegroups.com
Thank you, Maurício!!
I will give it a try today. Does it output the threshold values?
Hugo
To unsubscribe from this group and stop receiving emails from it, send an email to lavaan+unsubscribe@googlegroups.com.
To post to this group, send email to lav...@googlegroups.com.
Visit this group at https://groups.google.com/group/lavaan.
For more options, visit https://groups.google.com/d/optout.
Hugo Harada
Jul 19, 2018, 1:26:04 PM7/19/18
to lav...@googlegroups.com
Maurício,
When I run the EFA for only the 5 items for factor 1 asking for 2 factors, it gives me an error of negative variance in some cases (got the same error with the LSAT data).
Yes, i could confirm what you noted. And I can see the threshold values. ;-)
Feels like I need more items per factor. See equations (2.6) e (2.7) in the pdf attached. I will try with another data and return.
Thank you.
Hugo
Hugo Harada
Jul 19, 2018, 10:34:52 PM7/19/18
to lav...@googlegroups.com
Mauricio, Terence.
I have ran another dataset using package semtools. In this new dataset, I have 84 binary indicators. The first 39 are answers to portuguese tests and the remaining 45 are answers to a math test. There are 738 respondents. It is expected that the portuguese items should be mapped to a verbal factor, while the math items should go to mathematical factor. The problem of an unidentifiable model should be gone but it persists.
Warning message:In lav_model_vcov(lavmodel = lavmodel, lavsamplestats = lavsamplestats, :lavaan WARNING:
The variance-covariance matrix of the estimated parameters (vcov)does not appear to be positive definite! The smallest eigenvalue(= -1.552743e-02) is smaller than zero. This may be a symptom that
the model is not identified.
Any ideas? Code used is as below. The data is also attached.
rm(list=ls())library(semTools)mydata = read.csv("EQUIPE-2017-S1-PROVA034-QUIZ538-DICOTOMICA.TXT")mydata.factor <- mydatamydata.factor[,2:176]<- lapply(mydata.factor[,2:176],as.integer)
# language and maths exams onlyLC.MT = cbind(mydata.factor[,47:61],mydata.factor[,63:86],mydata.factor[,132:176])
### run EFA with WLSMV
ef2_irt <- efaUnrotate(data=LC.MT,estimator="wlsmv",nf=2,start=FALSE,std.lv=TRUE,ordered =colnames(LC.MT),debug=TRUE)
summary(ef2_irt, std = TRUE)inspect(ef2_irt, "std")## use oblique rotationef2_ob <- oblqRotate(ef2_irt)summary(ef2_ob,suppress=.001)
Regards,
Hugo
Mauricio Garnier-Villarreal
Jul 20, 2018, 12:40:51 AM7/20/18
to lavaan
Hugo
You dont need the std.lv=T, the EFA function sets the identification constraints needed for estimation.
When I run this I dont get an error
ordered =colnames(LC.MT))
summary(ef2_irt, std = TRUE)
Could it be that you dont have the latest version of semTools or lavaan?
This works for me with this session and package versions
> sessionInfo() R version 3.4.4 (2018-03-15) Platform: x86_64-pc-linux-gnu (64-bit) Running under: Ubuntu 18.04 LTS Matrix products: default BLAS: /usr/lib/x86_64-linux-gnu/openblas/libblas.so.3 LAPACK: /usr/lib/x86_64-linux-gnu/libopenblasp-r0.2.20.so locale: [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8 LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8 [7] LC_PAPER=en_US.UTF-8 LC_NAME=C LC_ADDRESS=C LC_TELEPHONE=C LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C attached base packages: [1] stats graphics grDevices utils datasets methods base other attached packages: [1] GPArotation_2014.11-1 semTools_0.5-0 lavaan_0.6-2.1271 loaded via a namespace (and not attached): [1] MASS_7.3-50 compiler_3.4.4 tools_3.4.4 mnormt_1.5-5 pbivnorm_0.6.0 stats4_3.4.4
Hugo Harada
Sócio-fundador - COOAdaptativa Inteligência Educacional S.A.
Trab: (11) 3052-3117 / Cel: (11) 96345-0390
Rua Claudio Soares, 72 - Sala 411Pinheiros, CEP 05422-030, São Paulo - SP
http://www.adaptativa.com.br
Hugo Harada
Jul 20, 2018, 9:11:24 AM7/20/18
to lav...@googlegroups.com
Mauricio,
Thank you for your reply.
Hmmm.... This is what I get when I run the code above...
Warning message:In lav_model_vcov(lavmodel = lavmodel, lavsamplestats = lavsamplestats, :lavaan WARNING:The variance-covariance matrix of the estimated parameters (vcov)does not appear to be positive definite! The smallest eigenvalue(= -1.552743e-02) is smaller than zero. This may be a symptom thatthe model is not identified.
And the factor values output are clearly not right...
Latent Variables:Estimate Std.Err z-value P(>|z|) Std.lv Std.all
factor1 =~LC051 (l1_1) 0.000 4482.551 0.000 1.000 0.000 0.000LC052 (l2_1) 0.000 4356.696 0.000 1.000 0.000 0.000LC053 (l3_1) -0.000 3910.838 -0.000 1.000 -0.000 -0.000LC054 (l4_1) -0.000 3925.346 -0.000 1.000 -0.000 -0.000
As for sessioninfo(), the only difference seems to be the R version and the OS. I will try to run my code in a linux virtual machine and try to confirm this is a platform dependent problem. In the meantime, could you send me the output of
summary(ef2_irt, std = TRUE)?
R version 3.5.1 (2018-07-02)Platform: x86_64-w64-mingw32/x64 (64-bit)Running under: Windows 7 x64 (build 7601) Service Pack 1Matrix products: defaultlocale:[1] LC_COLLATE=Portuguese_Brazil.1252 LC_CTYPE=Portuguese_Brazil.1252 LC_MONETARY=Portuguese_Brazil.1252[4] LC_NUMERIC=C LC_TIME=Portuguese_Brazil.1252
attached base packages:[1] stats graphics grDevices utils datasets methods baseother attached packages:[1] GPArotation_2014.11-1 semTools_0.5-0 lavaan_0.6-2
loaded via a namespace (and not attached):
[1] MASS_7.3-50 compiler_3.5.1 tools_3.5.1 mnormt_1.5-5 pbivnorm_0.6.0 stats4_3.5.1
Thank you for your help.
Hugo
To unsubscribe from this group and stop receiving emails from it, send an email to lavaan+unsubscribe@googlegroups.com.
To post to this group, send email to lav...@googlegroups.com.
Visit this group at https://groups.google.com/group/lavaan.
For more options, visit https://groups.google.com/d/optout.
Yves Rosseel
Jul 20, 2018, 9:21:45 AM7/20/18
to lav...@googlegroups.com
> Warning message:
> In lav_model_vcov(lavmodel = lavmodel, lavsamplestats =
> lavsamplestats, :
> lavaan WARNING:
> The variance-covariance matrix of the estimated parameters (vcov)
> does not appear to be positive definite! The smallest eigenvalue
> (= -1.552743e-02) is smaller than zero. This may be a symptom that
> the model is not identified.
This warning is new in 0.6-2!
> In lav_model_vcov(lavmodel = lavmodel, lavsamplestats =
> lavsamplestats, :
> lavaan WARNING:
> The variance-covariance matrix of the estimated parameters (vcov)
> does not appear to be positive definite! The smallest eigenvalue
> (= -1.552743e-02) is smaller than zero. This may be a symptom that
> the model is not identified.
I am not sure how efaUnrotate() works, but if I want to run an EFA using
lavaan, I do three things:
1) std.lv = TRUE)
2) orthogonal = TRUE
3) use an 'echelon' pattern for the lambda matrix
for example, if you have 8 observed variables, and 6 factors, the lambda
structure would look like this:
x 0 0 0 0 0
x x 0 0 0 0
x x x 0 0 0
x x x x 0 0
x x x x x 0
x x x x x x
x x x x x x
x x x x x x
where 'x' is free, and 0 is fixed to zero.
This works for the continuous case. Not sure about the categorical case.
Yves.
Hugo Harada
Jul 20, 2018, 10:35:29 PM7/20/18
to lav...@googlegroups.com
Hi, Mauricio!
As for sessioninfo(), the only difference
seems to be the R version and the OS. I will try to run my code in a
linux virtual machine and try to confirm this is a platform dependent
problem. In the meantime, could you send me the output of
summary(ef2_irt, std = TRUE)?
I have built a new setup in a virtual machine but I still get the same results. R is 3.4.4. on Ubuntu now. It seems that the problem is not platform dependent. It is so frustrating not to be able to reproduce your results...
> sessionInfo()
R version 3.4.4 (2018-03-15)
Platform: x86_64-pc-linux-gnu (64-bit)
R version 3.4.4 (2018-03-15)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 16.04.4 LTS
Matrix products: default
BLAS: /usr/lib/libblas/libblas.so.3.6.0
LAPACK: /usr/lib/lapack/liblapack.so.3.6.0
locale:
[1] LC_CTYPE=pt_BR.UTF-8 LC_NUMERIC=C LC_TIME=pt_BR.UTF-8
[4] LC_COLLATE=en_US.UTF-8 LC_MONETARY=pt_BR.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=pt_BR.UTF-8 LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=pt_BR.UTF-8 LC_IDENTIFICATION=C
Matrix products: default
BLAS: /usr/lib/libblas/libblas.so.3.6.0
LAPACK: /usr/lib/lapack/liblapack.so.3.6.0
locale:
[1] LC_CTYPE=pt_BR.UTF-8 LC_NUMERIC=C LC_TIME=pt_BR.UTF-8
[4] LC_COLLATE=en_US.UTF-8 LC_MONETARY=pt_BR.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=pt_BR.UTF-8 LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=pt_BR.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] semTools_0.5-0 lavaan_0.6-2
loaded via a namespace (and not attached):
[1] MASS_7.3-50 compiler_3.4.4 tools_3.4.4 mnormt_1.5-5 pbivnorm_0.6.0 stats4_3.4.4
Regards,
Hugo
Hugo Harada
Jul 20, 2018, 10:56:01 PM7/20/18
to lav...@googlegroups.com
Hi, Yves!!
3) use an 'echelon' pattern for the lambda matrixfor example, if you have 8 observed variables, and 6 factors, the lambda structure would look like this:
Could you elaborate on how I can pass this information to lavaan? In lavaan syntax, we write common factors as a function of the observed variables, correct? Like, F1 =~ l1*item1... If I understood you correctly, you are suggesting that I should write the measurement model the other way around, correct?
X_j = lambda_ji * F_i with j=1..8 and i=1..6, where lambda_ji obbeys the echelon pattern..
How do I do that in lavaan syntax?
Regards,
Hugo
--
You received this message because you are subscribed to the Google Groups "lavaan" group.
To unsubscribe from this group and stop receiving emails from it, send an email to lavaan+unsubscribe@googlegroups.com.
To post to this group, send email to lav...@googlegroups.com.
Visit this group at https://groups.google.com/group/lavaan.
For more options, visit https://groups.google.com/d/optout.
Terrence Jorgensen
Jul 21, 2018, 8:39:29 AM7/21/18
to lavaan
Hello all,
Converged but with a warning about NPD asymptotic covariance matrix of model parameters, and all loadings were zero with huge SEs. I also noticed that both the y* Scales (~*~) and the residual variances were all fixed to 1 under the default parameterization = "delta", when only the scales should be 1 and the residuals should be 1 minus the variance explained by factors. Of course, with factor loadings == 0, then this is internally consistent. But it made me curious to try setting parameterization = "theta".
I don't have access to Hugo's data, but I found the same issue with the "datCat" data used for the semTools::measurementInvarianceCat() function's help page.
fit.delta <- efaUnrotate(datCat[1:8], nf = 2, start=FALSE)
summary(fit.delta)fit.theta <- efaUnrotate(datCat[1:8], nf = 2, start=FALSE, parameterization = "theta")
summary(fit.theta)
This seems to have solved whatever the problem was, because the factor loadings are estimated and the fixed scaling parameters differ from 1 (while the residual variances are fixed to 1).
I can't figure out why this problem occurs with , and I'm not sure whether it happened with previous versions of lavaan and/or semTools. Internally, semTools only writes syntax for the factor loadings and factor (co)variances, leaving the cfa() function to specify the residual variances, intercepts, thresholds, and/or scaling parameters as necessary for the data on hand. If I can't figure it out, I will just add a warning that users with categorical data should try setting parameterization = "theta".
The ?efaUnrotate help page states the identification constraints in the Details section, including that the dot products of any pairs of columns in the factor loading matrix are constrained to zero. I don't have access to the Johnson & Wichern (2002) book he cites, so I don't know why this particular constraint is necessary. But if you want to use Yves' echelon pattern, you can just write the syntax to do so. It is a simple pattern, so using the paste() function is straight-forward.
varNames <- colnames(datCat)[1:8]
nf <- 3 ## number of factors
syntax <- character(0)
for (i in seq_along(varNames)) {
syntax <- c(syntax, paste0("fac_", i, " =~ ", paste(varNames[i:length(varNames)], collapse = " + ")))
if (i == nf) break
}
syntaxTerrence Jorgensen
Jul 21, 2018, 9:15:34 AM7/21/18
to lavaan
FYI, I also just tried the default parameterization = "delta", but also set start = TRUE, and my example returns nonzero factor loadings (and residual variances therefore differ from 1) without a warning about NPD covariance matrix of model parameters:
fit.delta.start <- efaUnrotate(datCat[1:8], nf = 2, start=TRUE)
summary(fit.delta.start)This means the starting values from the factanal() function allows the optimizer to find a better solution than using lavaan's default starting values (i.e., the ones it uses when users provide none). I am stymied, but at least we know it can still work. I will add a warning for this specific scenario, with instructions for how users can fix it (either change parameterization= or set start = TRUE).
Mauricio Garnier-Villarreal
Jul 21, 2018, 8:15:38 PM7/21/18
to lavaan
I try delta and theta parameterization with Hugos data, I get the warning for both
Warning message: In lav_model_vcov(lavmodel = lavmodel, lavsamplestats = lavsamplestats, : |
lavaan WARNING: The variance-covariance matrix of the estimated parameters (vcov) does not appear to be positive definite! The smallest eigenvalue (= -7.495555e+01) is smaller than zero. This may be a symptom that the model is not identified. > ef2_irt <- efaUnrotate(data=LC.MT, + estimator="wlsmv", + nf=2, + start=F, + ordered =colnames(LC.MT), + parameterization="delta") |
Warning message: In lav_model_vcov(lavmodel = lavmodel, lavsamplestats = lavsamplestats, : |
lavaan WARNING:
The variance-covariance matrix of the estimated parameters (vcov)
does not appear to be positive definite! The smallest eigenvalue
(= -1.020010e-02) is smaller than zero. This may be a symptom that
the model is not identified.
| |
|
With simulated data I get that error with delta, but not theta. With delta all factor loadings are 0, with theta the factor loadings are not 0.
Here is the code I used to simulate binary data from IRT parameters, with 4 latent factors
library(MASS)
library(rockchalk)
###Generate person parameters
## subjects
N <- 1000
#theta <- rnorm(N,0,1)
theta <- mvrnorm(n=N, mu=c(0,0,0,0), Sigma = lazyCor(0.4, 4) )
head(theta)
cor(theta)
summary(theta)
var(theta)
###Generate item parameters
item.par <- list()
p <- list()
x <- list()
for(k in 1:4){
n <- 7
b <- rnorm(n,0,1)
a <- runif(n,0.5,3)
c <- runif(n,0,0.3)#rep(0,n)#
item.par[[k]] <- cbind(a,b,c)
###Create arrays for prob (p) and simulated data (x)
p[[k]] <- matrix(0,N,n)
x[[k]] <- matrix(0,N,n)
###Use a stochastic process to generate model-fitting data
for (i in 1:N) {
for (j in 1:n){
p[[k]][i,j] <- c[j] + (1-c[j])/(1+exp(-a[j]*(theta[i,k]-b[j])))
r <- runif(1,0,1)
if (r <= p[[k]][i,j]) {
x[[k]][i,j] <- 1
}
}
}
}
head(x[[2]])
## combine items from the 4 factors
x2 <- data.frame(x[[1]], x[[2]], x[[3]], x[[4]])
head(x2)
### run EFA with WLSMV
ef2_irt <- efaUnrotate(data=x2,
estimator="wlsmv",
nf=2,
start=F,
ordered =colnames(x2),
parameterization="delta")
summary(ef2_irt, std = TRUE)
inspect(ef2_irt, "std")
## use oblique rotation
ef2_ob <- oblqRotate(ef2_irt)
summary(ef2_ob,suppress=.001)
### run EFA with WLSMV: Theta
ef2_irt_theta <- efaUnrotate(data=x2,
estimator="wlsmv",
nf=2,
start=F,
ordered =colnames(x2),
parameterization="theta")
summary(ef2_irt_theta, std = TRUE)
inspect(ef2_irt_theta, "std")
## use oblique rotation
ef2_ob_theta <- oblqRotate(ef2_irt_theta)
summary(ef2_ob_theta,suppress=.001)
Mauricio Garnier-Villarreal
Jul 21, 2018, 8:17:00 PM7/21/18
to lavaan
With the simulated data, delta works with start=TRUE
Hugo Harada
Jul 21, 2018, 11:06:39 PM7/21/18
to lav...@googlegroups.com
Hello all,
I tried four scenarios with
start = TRUE | FALSE
parametrization = "theta" | "delta"
and I got different warnings depending on the START value regardless of the parametrization . "(vcov) does not appear to be positive definite" when START=FALSE, and "The starting values from the factanal function cannot be calculated." when START=TRUE.
My code and data are attached.
=================================================================================
+ start=TRUE,
+ parameterization = "delta",
+ ordered =colnames(LC.MT)
+ )
Error in efaUnrotate(data = LC.MT, estimator = "wlsmv", nf = 2, start = TRUE, :
The starting values from the factanal function cannot be calculated. Please use start = FALSE instead.
In addition: Warning message:
In log(e) : NaNs produced
=================================================================================
+ start=TRUE,
+ parameterization = "theta",
+ ordered =colnames(LC.MT)
+ )
Error in efaUnrotate(data = LC.MT, estimator = "wlsmv", nf = 2, start = TRUE, :
The starting values from the factanal function cannot be calculated. Please use start = FALSE instead.
In addition: Warning message:
In log(e) : NaNs produced
=================================================================================
parameterization = "theta",
ordered =colnames(LC.MT)
)
Warning message:
In lav_model_vcov(lavmodel = lavmodel, lavsamplestats = lavsamplestats, :
lavaan WARNING:
The variance-covariance matrix of the estimated parameters (vcov)
does not appear to be positive definite! The smallest eigenvalue
(= -1.209680e+02) is smaller than zero. This may be a symptom that
the model is not identified.
=================================================================================
### run EFA with WLSMV for ordered items
parameterization = "delta",
ordered =colnames(LC.MT)
)
Warning message:
In lav_model_vcov(lavmodel = lavmodel, lavsamplestats = lavsamplestats, :
lavaan WARNING:
The variance-covariance matrix of the estimated parameters (vcov)
does not appear to be positive definite! The smallest eigenvalue
(= -1.552743e-02) is smaller than zero. This may be a symptom that
the model is not identified.
=================================================================================
Terence,
The ?efaUnrotate help page states the identification constraints in the Details section, including that the dot products of any pairs of columns in the factor loading matrix are constrained to zero. I don't have access to the Johnson & Wichern (2002) book he cites, so I don't know why this particular constraint is necessary. But if you want to use Yves' echelon pattern, you can just write the syntax to do so. It is a simple pattern, so using the paste() function is straight-forward.
Thank you for the code!! I will give it a try.
Regards,
Hugo
--
You received this message because you are subscribed to the Google Groups "lavaan" group.
To unsubscribe from this group and stop receiving emails from it, send an email to lavaan+unsubscribe@googlegroups.com.
To post to this group, send email to lav...@googlegroups.com.
Visit this group at https://groups.google.com/group/lavaan.
For more options, visit https://groups.google.com/d/optout.
Yves Rosseel
Jul 22, 2018, 6:41:36 AM7/22/18
to lav...@googlegroups.com
Just one comment: the fabin3 based starting values used by lavaan (for
factor loadings) assume a CFA model. It is very likely that they give
trouble when trying to fit an EFA model.
If you are using the sem/cfa/lavaan functions directly, you can set
start = "simple" to avoid them.
I need to check for this setting and generate better starting values. In
fact, I may write my own efa() function, as this has been on my TODO
list for almost 8 years now.
Can someone open an issue about this on github (just to remind me)? I
will have a look at this after my holidays (end of August).
Yves.
factor loadings) assume a CFA model. It is very likely that they give
trouble when trying to fit an EFA model.
If you are using the sem/cfa/lavaan functions directly, you can set
start = "simple" to avoid them.
I need to check for this setting and generate better starting values. In
fact, I may write my own efa() function, as this has been on my TODO
list for almost 8 years now.
Can someone open an issue about this on github (just to remind me)? I
will have a look at this after my holidays (end of August).
Yves.
Hugo Harada
Jul 23, 2018, 12:42:38 AM7/23/18
to lav...@googlegroups.com, yros...@gmail.com
--
You received this message because you are subscribed to the Google Groups "lavaan" group.
To unsubscribe from this group and stop receiving emails from it, send an email to lavaan+unsubscribe@googlegroups.com.
To post to this group, send email to lav...@googlegroups.com.
Visit this group at https://groups.google.com/group/lavaan.
For more options, visit https://groups.google.com/d/optout.
Terrence Jorgensen
Jul 24, 2018, 7:44:07 AM7/24/18
to lavaan
Hi Hugo,
On Sunday, July 22, 2018 at 12:41:36 PM UTC+2, Yves Rosseel wrote:
If you are using the sem/cfa/lavaan functions directly, you can set
start = "simple" to avoid them.
You can actually pass that (or any) argument to lavaan() via the efaUnrotate() in semTools, which is just a function that automatically writes lavaan syntax for an EFA model and fits it with lavaan. The object it returns is a lavaan object.
If setting start = "simple" does not solve your problem, then perhaps try fitting an EFA using the factanal() function that semTools uses to find starting values. Finding out why factanal() cannot find a solution could present a solution, or perhaps even a limitation of your data.
Hugo Harada
Jul 24, 2018, 9:43:35 PM7/24/18
to lav...@googlegroups.com
Terrence,
Thank you for the tip. I will give it a try.
Hugo
--
You received this message because you are subscribed to the Google Groups "lavaan" group.
To unsubscribe from this group and stop receiving emails from it, send an email to lavaan+unsubscribe@googlegroups.com.
To post to this group, send email to lav...@googlegroups.com.
Visit this group at https://groups.google.com/group/lavaan.
For more options, visit https://groups.google.com/d/optout.
Reply all
Reply to author
Forward
0 new messages

