Please, Why my GUI lose something
135 views
Skip to first unread message
小明
Jan 6, 2021, 4:07:58 AM1/6/21
to cmtk-users
https://user-images.githubusercontent.com/16069593/103741709-84b66700-5034-11eb-9d56-b42aacee96b1.png
this link is a picture of my command and interface
Sebastien Tourbier
Jan 6, 2021, 4:30:44 AM1/6/21
to cmtk-users
Hi,
Have you selected your BIDS dataset directory? If not, this is normal.
Just click on File -> Load BIDS Dataset...
Then select your BIDS dataset directory and click OK.
You should be all set.
Best
Sebastien
小明
Jan 7, 2021, 3:48:27 AM1/7/21
to cmtk-...@googlegroups.com
Hi,
I use data from https://openneuro.org/datasets/ds000007/versions/00001 and also data of my own, and check them through BIDS Validator, https://bids-standard.github.io/bids-validator/ , there was no error in my dataset, just some Warning.
But I even couldn't load BIDS dataset correctly.
Could you show me a dataset, then I can compare it with my dataset,
or, give me some advice from your experience.
Thanks,
Best,
wei
Sebastien Tourbier <sebastien...@gmail.com> 于2021年1月6日周三 下午5:30写道:
--
You received this message because you are subscribed to a topic in the Google Groups "cmtk-users" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cmtk-users/oSjqfjiTcmg/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cmtk-users+...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/cmtk-users/20dc4032-5438-4946-ad62-36f799b3afc6n%40googlegroups.com.
祝好!
盛巍
Sebastien Tourbier
Jan 11, 2021, 3:43:09 AM1/11/21
to cmtk-users
Hi Wei,
Sorry for the delay, just back from a few days off.
This is strange. I will give a try on my side with https://openneuro.org/datasets/ds000007/versions/00001 and let you know as soon as I know more.
Best
Seb
Sebastien Tourbier
Jan 11, 2021, 5:30:46 AM1/11/21
to cmtk-users
By looking at DS000007, it contains anatomical T1w scans and task fMRI.
However, CMP3 is handling ONLY resting-state fMRI. So even if the dataset is BIDS compliant, it cannot be processed by CMP3.
Regarding your in-house dataset, is it task or resting-state fMRI?
Sebastien Tourbier
Jan 11, 2021, 5:47:10 AM1/11/21
to cmtk-users
I do not know which method you used to download the dataset but even if CMP3 is not designed to process task fMRI data, I still tried to download DS000007 from OpenNeuro using the AWS client and it loads without any issue on my side.
See screenshot below:
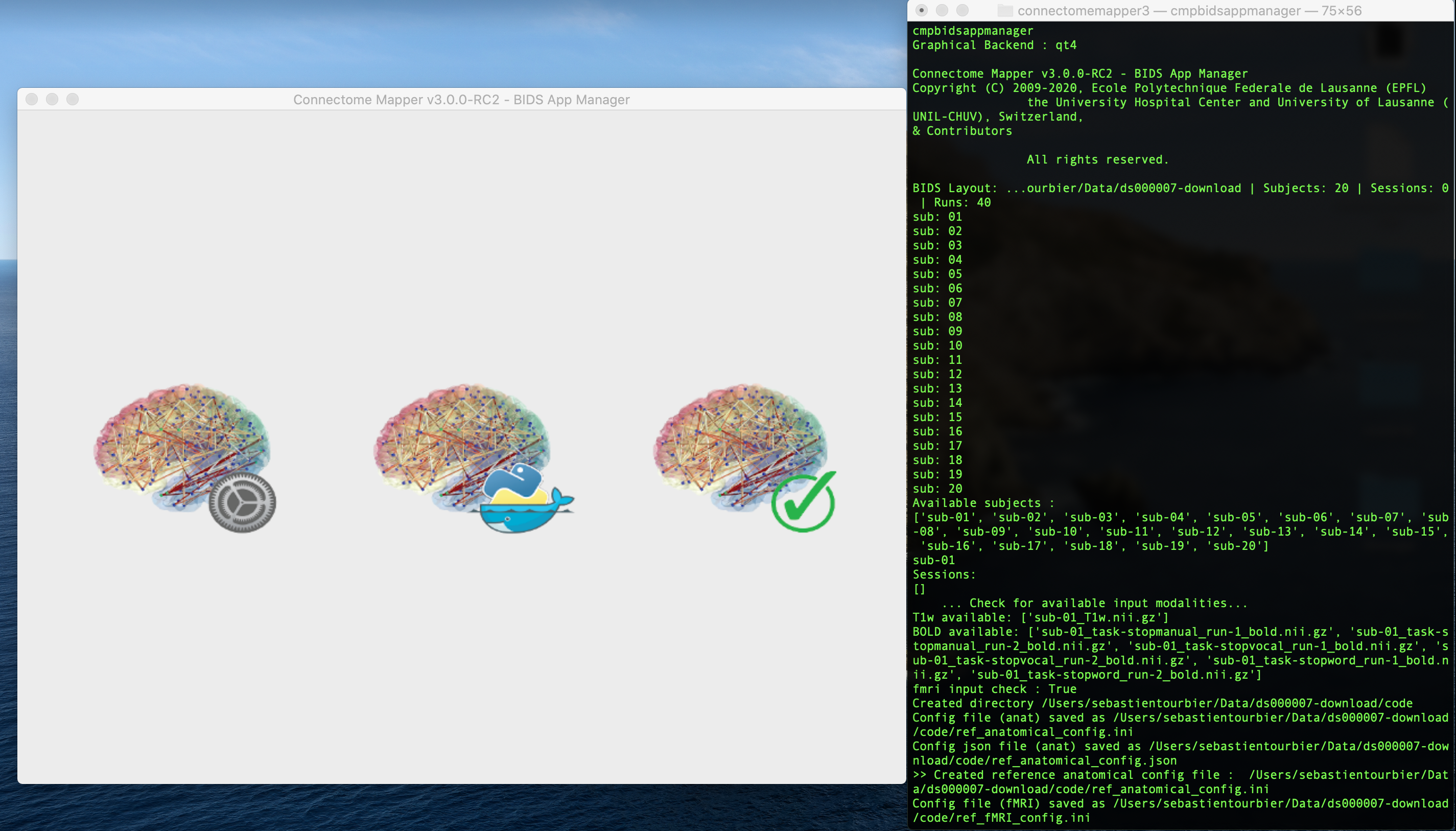
Note that I also tried to download this dataset using datalad. While the datalad install .... succeeded, the `datalad get .` command which download all the files failed (as you can see in the screenshot below).
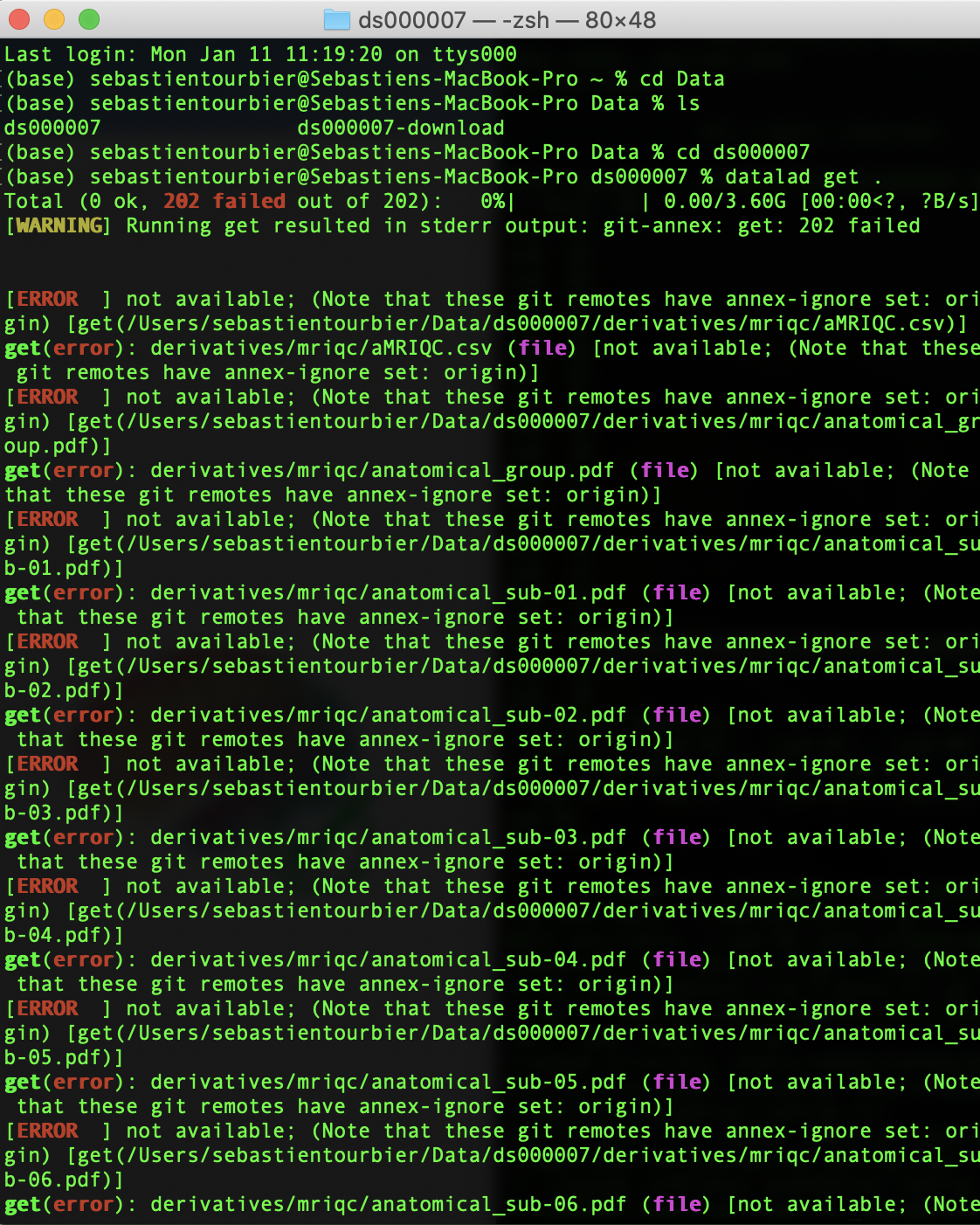
Best,
Sebastien
Sebastien Tourbier
Jan 11, 2021, 5:51:17 AM1/11/21
to cmtk-users
Also please find the sample dataset with sMRI, dMRI, and rsfMRI data that we use to test CMP3 on Zenodo at the following URL:
小明
Jan 13, 2021, 3:31:34 AM1/13/21
to cmtk-users
Hi, Sebastien , 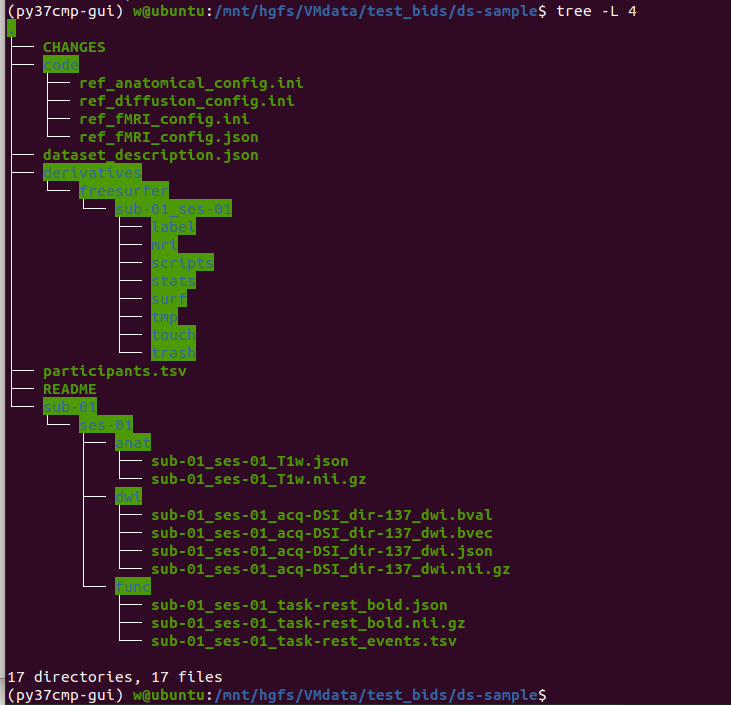
Thank you very much for your reply and tried a lot for me.
Now, I and load data correctly with my own data and your data from https://doi.org/10.5281/zenodo.3708962
The reason for the previous error had nothing to do with the data,
but with the installation of the software,
but I don't know what the problem is. I just reinstallation it.
Then, compared to https://connectome-mapper-3.readthedocs.io/en/latest/bidsappmanager.html
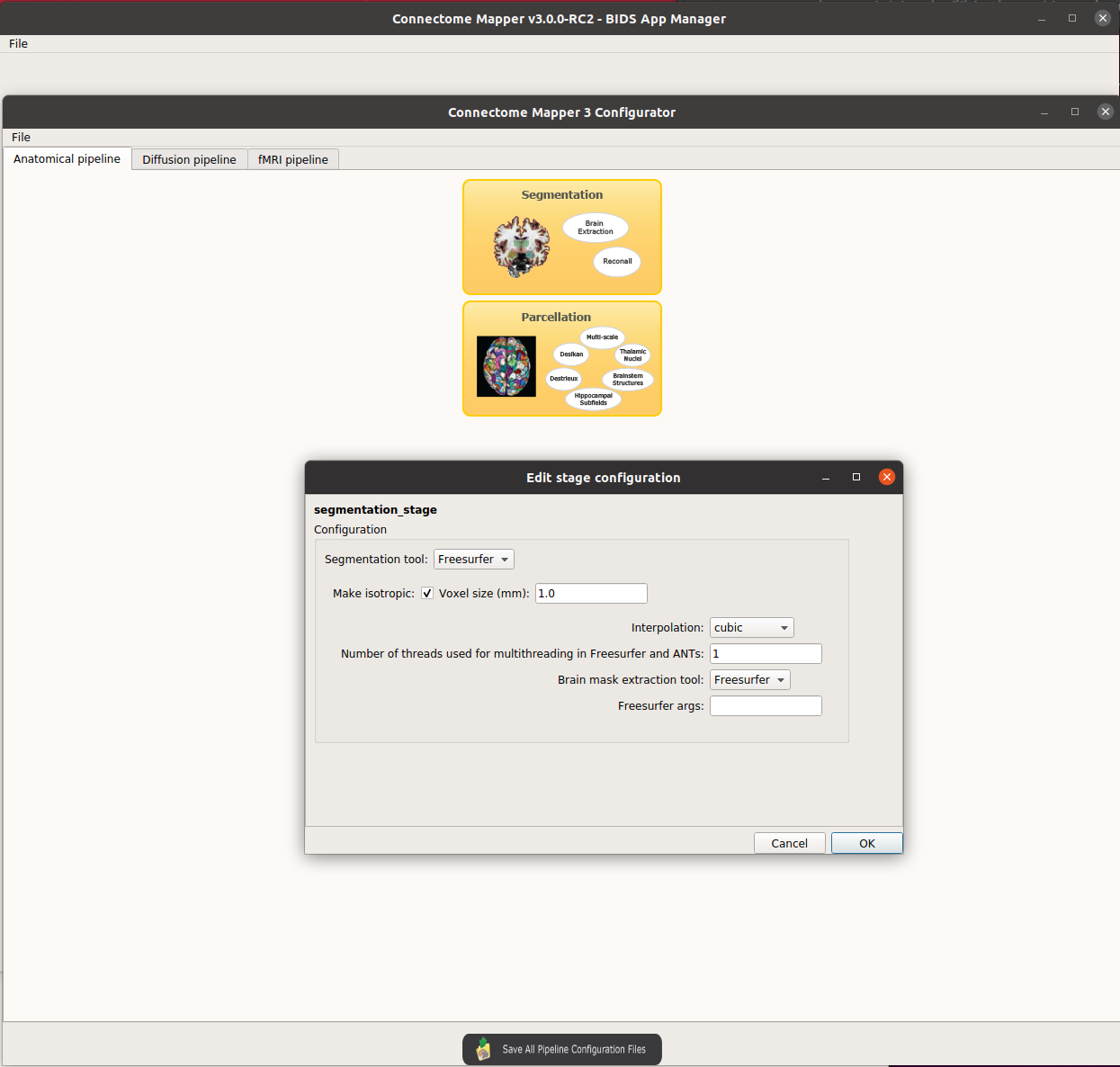
There are something like different, for example:
- Pipeline stage configuration -- Anatomical pipeline stages -- Segmentation : lose "Use existing freesurfer data " ,

GUI :

Best,
wei
小明
Jan 13, 2021, 3:54:31 AM1/13/21
to cmtk-users
Hi,
My intention is to use Lausanne multi-resolution atlas to reconstruction network (Creating the Connectome), use DTI data.
I know it's inappropriate, but I still want to ask if you have a better suggestion, for, other steps, I can processing them use other tools which I'm more familiar.
Before this, I have tried and got the 5 scales mgz files
- lausanne2008.scale1+aseg.mgz
- lausanne2008.scale2+aseg.mgz
- lausanne2008.scale3+aseg.mgz
- lausanne2008.scale4+aseg.mgz
- lausanne2008.scale5+aseg.mgz
Limited by my code ability, I couldn't get the key code about Creating the Connectome from cmp
Best,
wei
Sebastien Tourbier
Jan 13, 2021, 4:48:32 AM1/13/21
to cmtk-users
Hi Wei!
Great to hear you managed to go forward!
Indeed there are still some screenshots in the documentation that need to be updated so do not worry about that.
Actually, the first and second versions of CMP needed to select explicitly an existing Freesurfer directory if the data was already processed by Freesurfer.
But, now with the adoption of (1) Nipype for workflow management and (2) BIDS for organization, the good news is that it is not anymore needed :)
If the outputs of freesurfer are found in the derivatives folder, it will use them and will not re-run the recon-all process.
In particular, it should follow the following hierarchy:
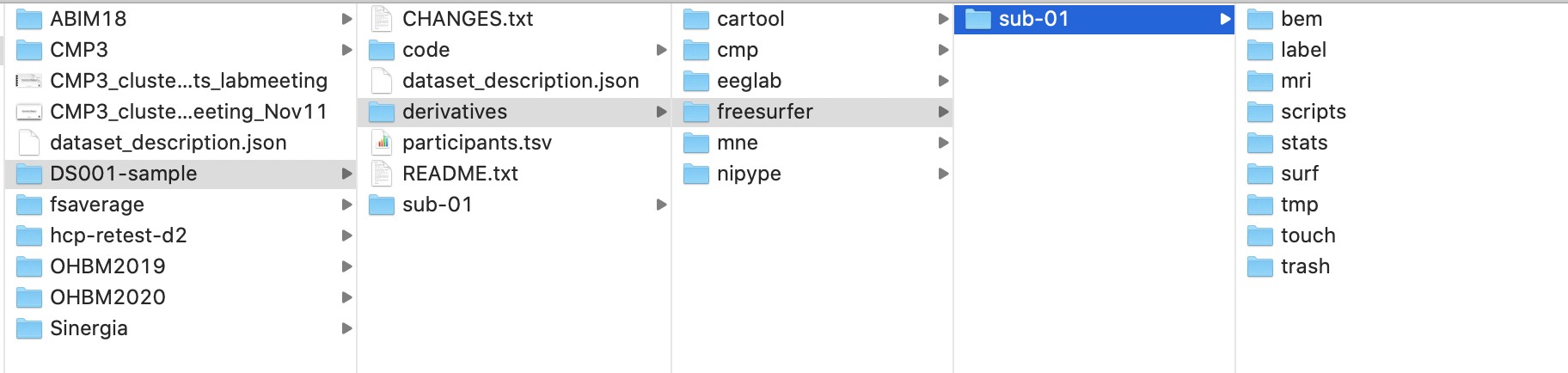
Sebastien Tourbier
Jan 13, 2021, 5:11:17 AM1/13/21
to cmtk-users
Hi Wei,
You can indeed perform all the steps (structural parcellation / diffusion preprocessing reconstruction and tractography / connectome creation) directly inside CMP3.
Parcellation
I would recommend to use Lausanne2018 that can generate ~4X faster and more accurately the original Lausanne2008 parcellation if not other extra structures are added i.e:
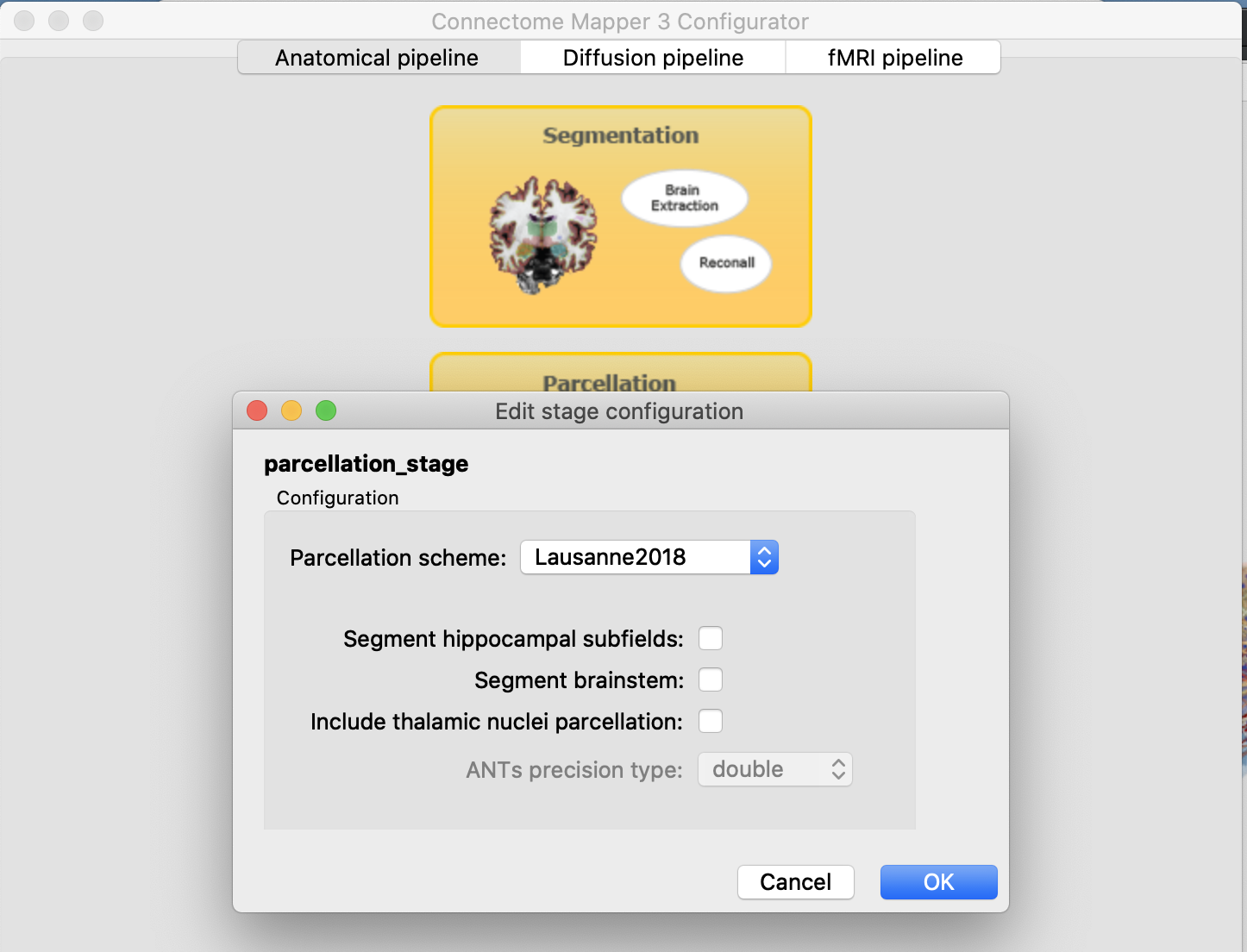
Diffusion preprocessing
I would first give a try with the following settings:
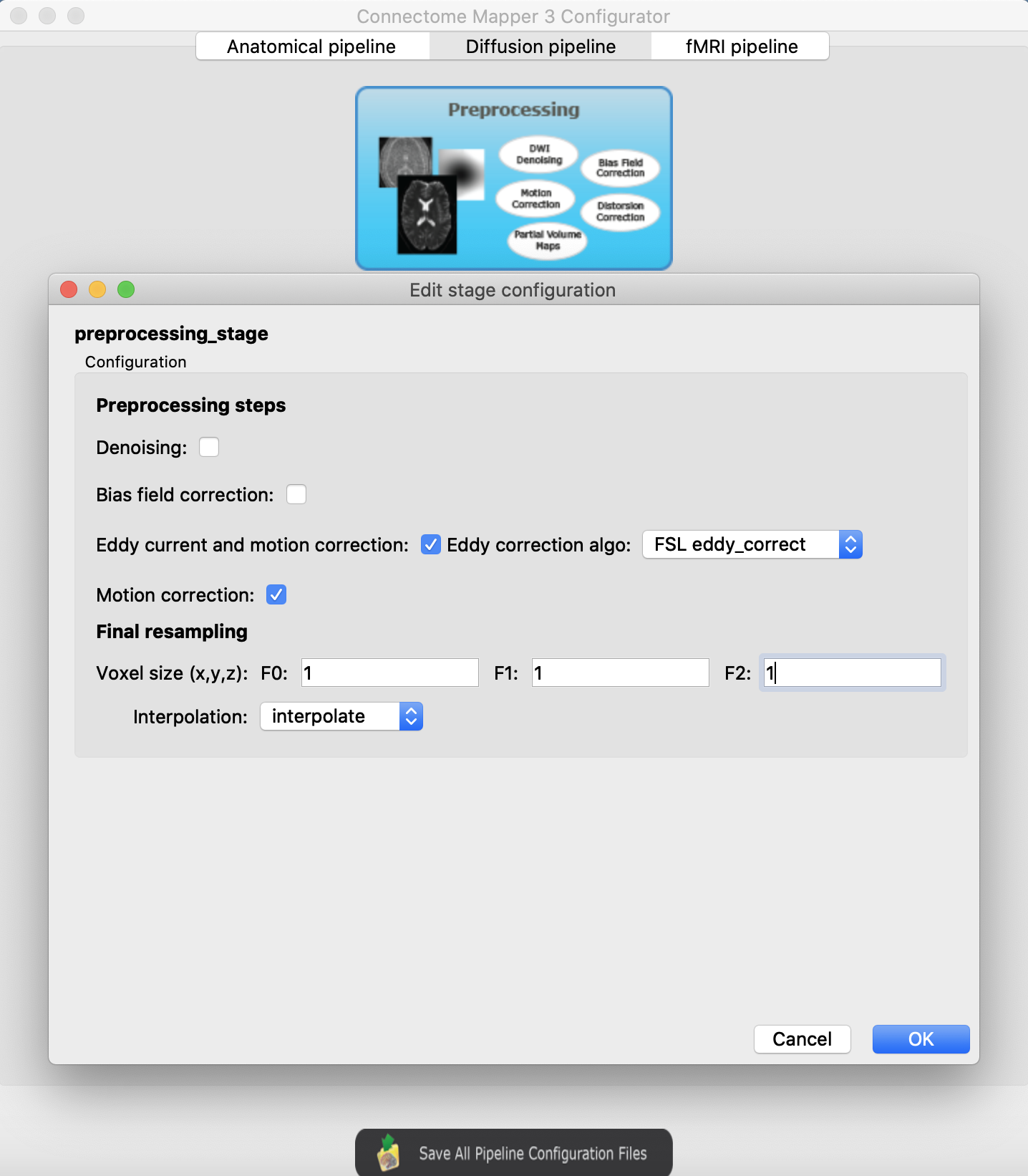
Diffusion reconstruction and tractography
In the case of DTI data, I would recommend you to use a Constrained Spherical Deconvolution model for estimating fiber orientation distribution functions which is able to solve crossing fibers in contrast to a basic tensor model. For instance, in our lab we process DTI data with 30 directions with a CSD order of 4.
Regarding the tractography strategy, I would start using the mrtrix deterministic method with seeding in the white-matter mask and a high number of desired streamlines (>2000000). Then, you can use SIFT to filter the streamlines and reduce false positive connections but be aware that it will result in a tractogram with 10% of the original fibers/streamlines (meaning that if you set # of streamlines to 10'000'000, the final tractogram after SIFT will consist of 1'000'000 streamlines)
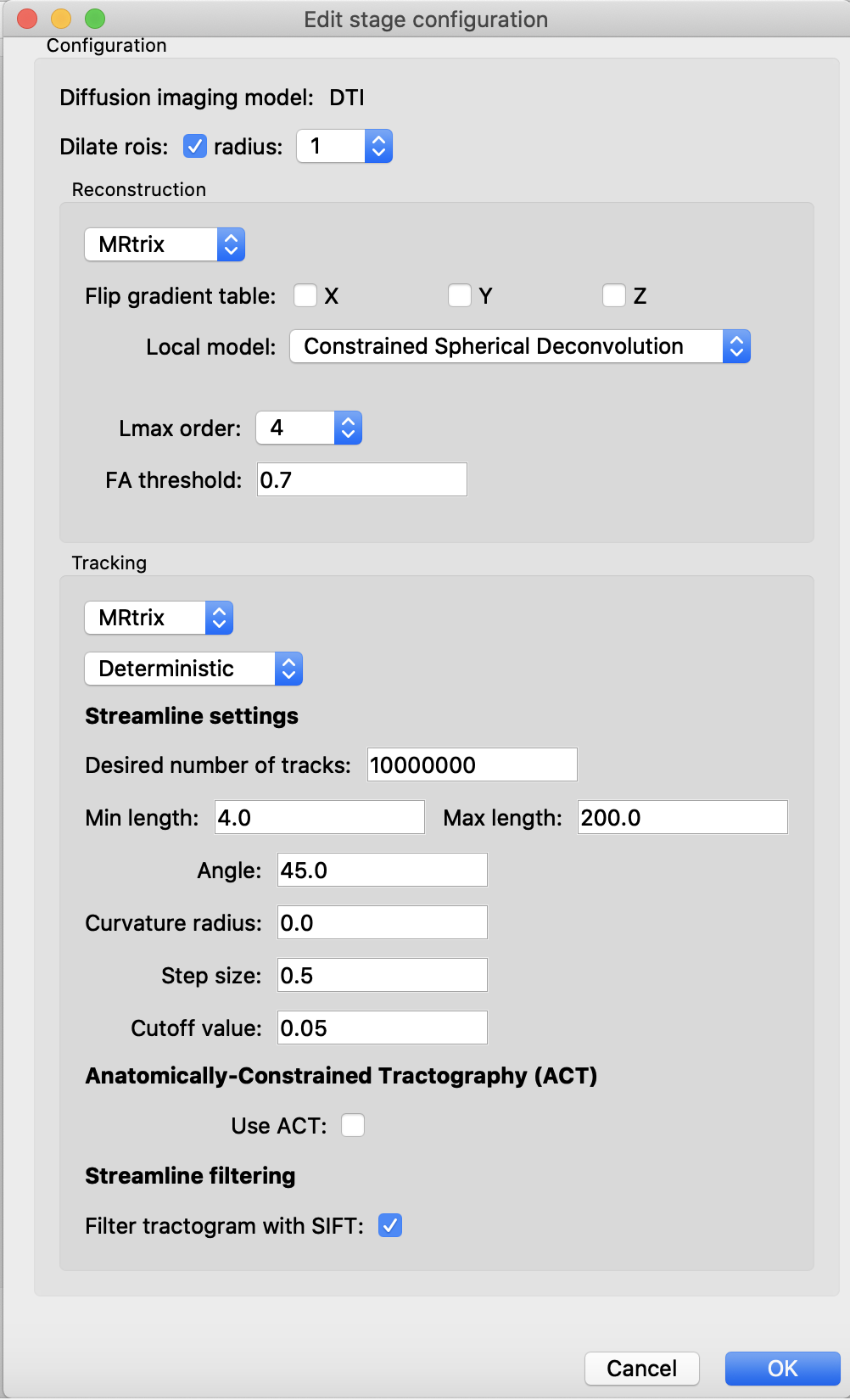
When all steps are configured, do not forget to save the configuration files.
All main ouputs (parcellation, processed dMRI, tractograms and connectomes) will be saved in derivatives/cmp/sub-01. See https://connectome-mapper-3.readthedocs.io/en/latest/outputs.html for more details.
In particular, you can find the description of parcellation nodes for each scale in their respective graphml files i.e.:

Best,
Sebastien
小明
Jan 13, 2021, 6:52:50 AM1/13/21
to cmtk-users
Hi Sebastien,
Thank you for your detailed reply and patient answer.
Best,
wei
小明
Jan 14, 2021, 6:13:03 AM1/14/21
to cmtk-users
Hi Sebastien,
There's something wrong here.
from ds-sample\derivatives\nipype\sub-01\ses-01\diffusion_pipeline\pypeline.log
210114-02:00:43,485 nipype.workflow INFO:
[Node] Running "dipy_mapmri" ("cmtklib.interfaces.dipy.MAPMRI")
210114-02:01:46,527 nipype.interface INFO:
Fitting MAP-MRI model
It run this step many hours, but no result.
then, from ds-sample\derivatives\cmp\sub-01\ses-01\sub-01_ses-01_log.txt
I find some Warning,
210114-02:01:46,527 nipype.interface INFO:
Fitting MAP-MRI model
/opt/conda/envs/py37cmp-core/lib/python3.7/site-packages/cvxpy/expressions/expression.py:550: UserWarning:
This use of ``*`` has resulted in matrix multiplication.
Using ``*`` for matrix multiplication has been deprecated since CVXPY 1.1.
Use ``*`` for matrix-scalar and vector-scalar multiplication.
Use ``@`` for matrix-matrix and matrix-vector multiplication.
Use ``multiply`` for elementwise multiplication.
warnings.warn(__STAR_MATMUL_WARNING__, UserWarning)
/opt/conda/envs/py37cmp-core/lib/python3.7/site-packages/cvxpy/expressions/expression.py:550: UserWarning:
This use of ``*`` has resulted in matrix multiplication.
Using ``*`` for matrix multiplication has been deprecated since CVXPY 1.1.
Use ``*`` for matrix-scalar and vector-scalar multiplication.
Use ``@`` for matrix-matrix and matrix-vector multiplication.
Use ``multiply`` for elementwise multiplication.
warnings.warn(__STAR_MATMUL_WARNING__, UserWarning)
/opt/conda/envs/py37cmp-core/lib/python3.7/site-packages/cvxpy/expressions/expression.py:550: UserWarning:
This use of ``*`` has resulted in matrix multiplication.
Using ``*`` for matrix multiplication has been deprecated since CVXPY 1.1.
Use ``*`` for matrix-scalar and vector-scalar multiplication.
Use ``@`` for matrix-matrix and matrix-vector multiplication.
Use ``multiply`` for elementwise multiplication.
warnings.warn(__STAR_MATMUL_WARNING__, UserWarning)
/opt/conda/envs/py37cmp-core/lib/python3.7/site-packages/dipy/reconst/mapmri.py:418: UserWarning: Optimization did not find a solution
warn('Optimization did not find a solution')
/opt/conda/envs/py37cmp-core/lib/python3.7/site-packages/dipy/reconst/mapmri.py:396: RuntimeWarning: divide by zero encountered in true_divide
data_norm = np.asarray(data / data[self.gtab.b0s_mask].mean())
/opt/conda/envs/py37cmp-core/lib/python3.7/site-packages/dipy/reconst/mapmri.py:396: RuntimeWarning: invalid value encountered in true_divide
data_norm = np.asarray(data / data[self.gtab.b0s_mask].mean())
/opt/conda/envs/py37cmp-core/lib/python3.7/site-packages/cvxpy/expressions/expression.py:550: UserWarning:
And this warning repeated many times, up to now.
I'm going to try again, then, I will report my results
I
在2021年1月13日星期三 UTC+8 下午6:11:17<Sebastien Tourbier> 写道:
Sebastien Tourbier
Jan 14, 2021, 8:16:00 AM1/14/21
to cmtk-...@googlegroups.com
Hi Wei,
As you said this is only warning and not errors, and it is coming directly from Dipy.
But MAP-MRI is only dedicated to multi-shell DWI and you SHOULD NOT use MAP-MRI if you have DTI data.
In the case of multi-shell data, MAP-MRI requires to set the small delta (diffusion time) and big delta (pulse separation time) which are specific to the multi-shell DWI acquisition.
Hope this helps
Sebastien
To view this discussion on the web visit https://groups.google.com/d/msgid/cmtk-users/73472fd5-b40c-478b-8f98-f96b9e9b8f4dn%40googlegroups.com.
Sebastien Tourbier
Jan 14, 2021, 8:17:46 AM1/14/21
to cmtk-...@googlegroups.com
Also please could you open new topics in the future, this would help others to access support information.
Thanks a lot in advance,
Sebastien
Reply all
Reply to author
Forward
0 new messages
