Re: [pflotran-users: 998] Distorted structured grid
Gautam Bisht
Thanks to everybody for the comments.
I know that a correction term is needed to take account of the tangential flux. At the same time when the mesh distortions are small, we can live with a first order approximation. I know you are working to implement the mimetic finite difference (MFD) for the MPHASE module, this should take care not only of unstructured meshes but also of structured distorted meshes. Is it still a priority?
We would like to test how the MFD already implemented for the RICHARDS mode.
This is implemented only for structured meshes, but it should handle distortions correctly, right?
Are the quadrilateral faces divided in the two triangular ones in the local MFD discretisation (i.e. is the hex considered as a general polyhedron and split into tets?)
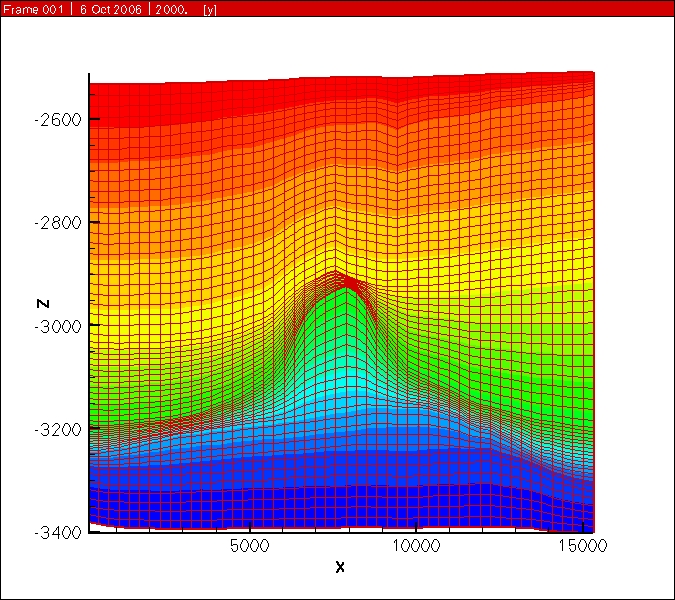
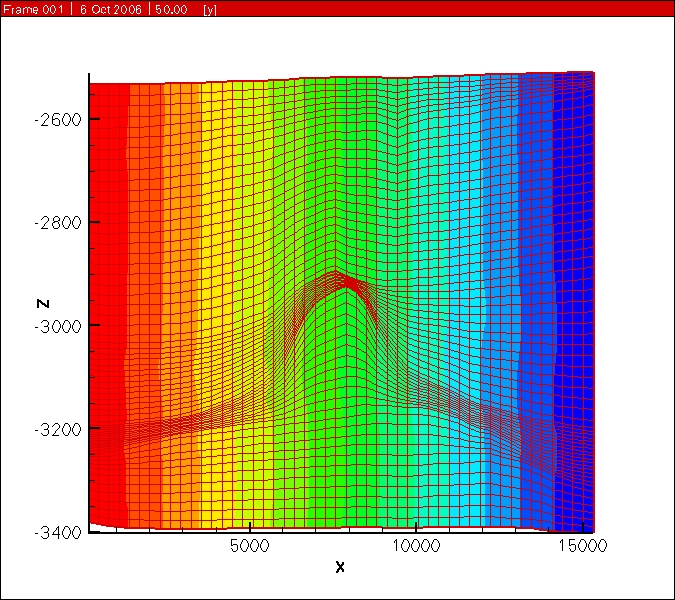
We came across several models where the use of distorted structured meshes can be very handy (i.e. representing the real geology but keeping the simplicity and efficiency of structured meshes). We are considering to implement a mesh loader for structured meshes to allow different vertical node distributions for each vertex column. From the code we analysed so far it seems that we need to introduce/change several functions called in the initialisation to accomplish two main tasks: (a) create new data structure to read in the additional dz,
(b) use some of the UMesh tools to compute cell centres, volume and areas.
However, once the grid is formed there should not be any other changes, right?
Do you see any major issues in doing this?
Will you be interested in checking and adding this extra feature to the standard release once we have done it? ... a more sophisticated way to handle fluxes can be added later
Paolo
On Sunday, September 8, 2013 8:47:45 PM UTC+2, Gautam Bisht wrote:Hi Paolo,On Sun, Sep 8, 2013 at 11:06 AM, Satish Karra <satk...@gmail.com> wrote:
Can you generate a voronoi mesh with this domain? If yes, you can use the explicit unstructured grid method of reading that voronoi mesh. The problem with orthogonality would be solved.--Satish
Sent from my iPhoneI think you would need to add some correction terms to get sufficiently accurate results....PeterOn Sep 8, 2013, at 1:44 AM, Paolo Orsini - BLOSint <paolo....@gmail.com> wrote:Hi,Is it possible to define a structured grid defining dx and dy spacing in the usual way, but having a dz spacing that varies for each column of nodes?ExampleSay we have a structured grid with NX x NY x NZ cells, (NX+1)x(NY+1)x(NZ+1) points.Is it possible to input NX dx in x direction, NY dy in y direction, and (NX+1)x(NY+1)XNZ dz spacing for the vertical direction Z? i.e. one each vertical node columnI believe for a structured grid, PFLOTRAN only allows specification of NZ 'dz' values [ not (NX+1)*(NY+1)*NZ dz values].Unstructured grid format in PFLOTRAN will be able to accommodate your grid. You can use:- Implicit unstructured grid format: in which you should split hexahedrons into prismatic control volumes, or- Explicit unstructured grid format: in which you can use hexahedron control volumes.-GautamI know that in this case the horizontal faces will not respect the orthogonality condition with respect to the line joining the cell centres, however in many reservoir problems the variation of Z are very small compared to the dx and dy spacing, and the error should be contained.If not implemented, it should not be a huge work to adapt the code to do this, or am I missing something?what do you think?Paolo
Paolo Orsini - BLOSint
Jed Brown
> - Using parallel unstructured requires a more complex and less efficient
> domain partition. The load balancing will be always easier to control and
> more efficient when using structured grids
expensive in some parts of the domain and load balancing with
non-uniform weights is trivial with unstructured grids, but tends to
break structured partitions.
> - When using the parallel unstructured grids, ParMetis is required, which
> is not free, and does not follow the open source model. One solution could
> be to replace Parmetis with the Scotch library, which from what I gathered
> has been already linked to PETSC. However, I don't know how much work there
> is to do and how hard it would be. And if there is any interest in doing
> this. Any comments on this?
option -mat_partitioning_type ptscotch.
Note that in our experience, PTScotch is often a bit higher quality than
ParMETIS, but tends to be substantially more expensive.
Gautam Bisht
Hi Gautam,
Thanks for your answers/comments.Our idea is to implement the "Standard FV with distorted cell without tangential flux correction" [not implemented for structured grid]
Paolo Orsini - BLOSint
Hammond, Glenn E
Note that you will need to read (nx+1)*(ny+1) dz’s. This is all based on the number of connections in the horizontal. A similar capability existed in the past with general_grid.F90, but that capability is out of date. It lists me as the author, but I simply converted routines that Lichtner or Lu wrote years earlier.
Glenn
--
You received this message because you are subscribed to the Google Groups "pflotran-dev" group.
To view this discussion on the web visit
https://groups.google.com/d/msgid/pflotran-dev/bc382b75-f412-4ff8-bd24-bf0954a0dac0%40googlegroups.com.
Gautam Bisht
To view this discussion on the web visit https://groups.google.com/d/msgid/pflotran-dev/DF0B2AE3F6166745843F1066C7E2F38E8307C7%40EXMB01.srn.sandia.gov.
Karra, Satish
As you said this can already be handled by PFLOTRAN using the unstructured grids, but we see the following problems/limitations:
- Using parallel unstructured requires a more complex and less efficient domain partition. The load balancing will be always easier to control and more efficient when using structured grids
- When using the parallel unstructured grids, ParMetis is required, which is not free, and does not follow the open source model. One solution could be to replace Parmetis with the Scotch library, which from what I gathered has been already linked to PETSC. However, I don't know how much work there is to do and how hard it would be. And if there is any interest in doing this. Any comments on this?
It basically seems a shame not to use structured meshes when is possible, and we realised that structured meshes are still heavily used by reservoir engineers.
A tangential flux correction to this distorted-structured grid could be added later, this requires more work and a more careful thought.
We want to test the MFD (as you recommended), and look into more classical approaches ( least square method, cell gradient reconstruction [e.g. Turner proposes several methods] ). Again, the structured nature of the grid should give more options for the implementation of a correction.
Having said that, I recognise the power of unstructured grids, and having a proper unstructured solver in PFLOTRAN for every mode will be great (e.g. MFD for unstructured meshes in every mode developed). At the same time, having also a structured grid capability that can handle real geometries at this stage can attract many more users.
What do you think about this distorted-structured alternative ?
I would argue that you can still use two-point flux on unstructured/distorted grids, just make sure that you generate a voronoi mesh for your domain. PFLOTRAN can read the areas of faces and volumes via explicit unstructured capability. This can handle real geometries and domains with discrete fracture networks (you can find a two intersecting fracture example in pflotran-dev/regression_tests/default/discretization/dfn_explicit.in) and give results with better accuracy (than a hex mesh, in your case).
Thanks,
Satish
Peter Lichtner
To view this discussion on the web visit https://groups.google.com/d/msgid/pflotran-dev/CE783CFB.483B%25satkarra%40lanl.gov.
Jed Brown
> Hi Jed,
>
> Thanks for this.
>
> So to use PTScotch, all we need to do is to (a) recompile PETSC (adding the
> option "--download-ptscotch -mat_partitioning_type ptscotch", and (b)
> change two PFLOTRAN guards from PETSC_HAVE_PARMETIS to PETSC_HAVE_SCOTCH
> (in discretization.F90 and unstructured_grid.F90) ?
Change the guards to accept either (assuming you want compile-time
failure when neither is available).
> That's all?
Should be, yes.
> Paolo
>
>
> On Mon, Oct 7, 2013 at 6:10 PM, Jed Brown <jedb...@mcs.anl.gov> wrote:
>
>> Paolo Orsini <paolo....@gmail.com> writes:
>> > When you say that PTScotch tends to be substantially more expensive, do
>> you
>> > refer to computational cost? Or actual financial cost?
>>
>> Computational cost.
>>
>> PTScotch uses the CeCILL-C V1 license, which is similar to LGPL.
>>
Paolo Orsini
Jed Brown
>
> The problem is that PFLOTRAN calls MatMeshToCellGraph to create the Dual
> graph, operation which is done by using the the parmetis function
> ParMETIS_V3_Mesh2Dual.
>
> Is there an equivalent function of MatMeshToCellGraph that uses PTScotch?
something similar that could be used. If they do, it would be worth
adding, but it's not currently supported.
Jed Brown
and so others can comment.
Paolo Orsini <paolo....@gmail.com> writes:
> I really don't know how the DualGraph is going to affect the the parallel
> computation efficiency...
> I am trying to use this partitioning as a black box, not sure what's the
> right thing to do..
>
> In the code calling the partitioning (PFLOTRAN), i can see that MPIAdj is
> built, then the Dual graph is computed, and finally the partitioning takes
> place, see below:
>
> !!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!
> call MatCreateMPIAdj(option%mycomm,num_cells_local_old, &
> unstructured_grid%num_vertices_global, &
> local_vertex_offset, &
> local_vertices,PETSC_NULL_INTEGER,Adj_mat,ierr)
>
> call MatMeshToCellGraph(Adj_mat,num_common_vertices,Dual_mat,ierr)
You can MatConvert Adj_mat to AIJ or assemble as AIJ to begin with, then
use MatMatTransposeMult to create Dual_mat (which will be in AIJ
format).
> .........
>
> call MatPartitioningCreate(option%mycomm,Part,ierr)
> call MatPartitioningSetAdjacency(Part,Dual_mat,ierr)
> call MatPartitioningSetFromOptions(Part,ierr)
> call MatPartitioningApply(Part,is_new,ierr)
> call MatPartitioningDestroy(Part,ierr)
>
> allocate(cell_counts(option%mycommsize))
> call ISPartitioningCount(is_new,option%mycommsize,cell_counts,ierr)
> num_cells_local_new = cell_counts(option%myrank+1)
> call
> MPI_Allreduce(num_cells_local_new,iflag,ONE_INTEGER_MPI,MPIU_INTEGER, &
> MPI_MIN,option%mycomm,ierr)
> !!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!
>
> You are telling me we can build a dual graph simply multiplying Adj_mat by
> its transpose?
Yeah, doesn't it connect all cells that share at least one vertex? That
is exactly the nonzero pattern of the matrix product.
> Why using a simple Aij when we are pre-processing a parallel process?
MPIAIJ implements MatMatTransposeMult, but we don't have an
implementation for MPIAdj (without calling ParMETIS).
> Sorry, may be what I want to do is less black box than thought...
>
> And thanks again for your patience and help
>
> Paolo
>
>
>
> On Fri, Nov 1, 2013 at 9:17 PM, Jed Brown <jedb...@mcs.anl.gov> wrote:
>
>> Paolo Orsini <paolo....@gmail.com> writes:
>>
>> > Hi Jed,
>> >
>> >
>> > Is it possible to do the partitioning with PTSchotch, without
>> > using MatMeshToCellGraph?
>> >
>> > Is there any other function (not using ParMetis) to compute the adjacency
>> > graph needed by MatPartitioningSetAdjacency, for parallel unstructured
>> > grids?
>>
>> You can either compute the dual graph yourself or we can implement that
>> function without calling ParMETIS. Actually, I believe the operation is
>> just symbolic
>>
>> DualGraph = E * E^T
>>
>> where E is the element connectivity matrix. So you could build a normal
>> (AIJ instead of MPIAdj) matrix with the connectivity, then call
>> MatMatTransposeMult().
>>
>> Would that work for you?
>>
Paolo Orsini
Jed Brown
> Multiplying the connectivity matrix by its tranpose returns an ncell x
> ncell matrix with non-zero entries only for those cell sharing at least one
> node. Hence the Dual Graph we need....
>
> Following your suggestions I change the code as follows:
>
> !!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!
> call MatCreateMPIAdj(option%mycomm,num_cells_local_old, &
> unstructured_grid%num_vertices_global, &
> local_vertex_offset, &
> local_vertices,PETSC_NULL_INTEGER,Adj_mat,ierr)
>
> call MatConvert(Adj_mat, MATAIJ,MAT_INITIAL_MATRIX,AIJ_mat,ierr)
>
> call
> MatMatTransposeMult(AIJ_mat,AIJ_mat,MAT_INITIAL_MATRIX,PETSC_DEFAULT_DOUBLE_PRECISION,Dual_mat,ierr)
> !! call MatMeshToCellGraph(Adj_mat,num_common_vertices,Dual_mat,ierr)
>
> call MatDestroy(Adj_mat,ierr)
> call MatDestroy(AIJ_mat,ierr)
> !!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!
>
> When I try to run, the MatConvert takes ages, (I suppose I can form
> directly the AIJ_mat),
certainly why it takes a long time. That's easy to fix in PETSc (at
least for AIJ).
PETSc devs, why doesn't it make one pass counting and another pass
inserting?
> the PTScotch partitioning seems quick, but then I get an error later
> on in the code, when attempting to compute the internal
> connection.... Something is wrong...
Paolo Orsini
Jed Brown
> The computation of the dual graph of the mesh is a bit more complicated
> than multiplying the adjacency matrix by its transpose, but not far off.
> With this operation, also the cells that share only one node are connected
> in the dual graph.
> Instead, the minimum number of common nodes is >1 (2 in 2D probelms, 3 in
> 3D problems). In fact, this is an input of MatMeshToCellGraph, I should
> have understood this before.
>
> This can be computed doing the transpose adjacency matrix (Adj_T), then
> doing the multiplication line by line of Adj time Adj_T, and discard the
> non zero entries coming from to elements that share a number of nodes less
> than the minimum number of common nodes imposed. I have not implement this
> yet, any suggestion is welcome.
the resulting matrix. It'll take a bit more memory, but probably not
prohibitively much.
> I also found out that Schotch has a facility to compute a dual graph from a
> mesh, but not PTScotch.
> Once the graph is computed, PTSchotch can load the central dual graph, and
> distribute it into several processors during the loading.
> Am i right to say that PETSC is interfaced only with PTSchotch and not with
> Scotch?
would not be scalable (memory or time).
> To check if the PTSchotch partition works (within PFLOTRAN ), I am
> computing a DualMat with parmetis, saving it into a file. Then I recompile
> the code (using a petsc compiled with ptscotch), an load the DualMat from a
> file rather then forming a new one. I did a successful test when running on
> one processor. but I am having trouble when try on more.
that the distribution matches what you expect). You'll have to be more
specific if you want more help.
> I though the the dual graph was computed only once, even during the mpi
> process, instead it seems to be recomputed more than once. Not sure why....
> sure i am missing something ???
figure out if you set a breakpoint in the debugger.
Gautam Bisht
Hi Jed,I found out what the problem is.First of all I printed all the matrices out, to be sure that MatConvert was working fine.And that was ok: AIJ_mat had the same connectivity table than Adj_mat.Then I compared the dual graph (stored in DualMat) computed by the Parmetis routine (MatMeshToCellGraph) and with MatMatTransposeMult. And they were quite different.The computation of the dual graph of the mesh is a bit more complicated than multiplying the adjacency matrix by its transpose, but not far off. With this operation, also the cells that share only one node are connected in the dual graph.Instead, the minimum number of common nodes is >1 (2 in 2D probelms, 3 in 3D problems). In fact, this is an input of MatMeshToCellGraph, I should have understood this before.This can be computed doing the transpose adjacency matrix (Adj_T), then doing the multiplication line by line of Adj time Adj_T, and discard the non zero entries coming from to elements that share a number of nodes less than the minimum number of common nodes imposed. I have not implement this yet, any suggestion is welcome.I also found out that Schotch has a facility to compute a dual graph from a mesh, but not PTScotch.Once the graph is computed, PTSchotch can load the central dual graph, and distribute it into several processors during the loading.Am i right to say that PETSC is interfaced only with PTSchotch and not with Scotch?To check if the PTSchotch partition works (within PFLOTRAN ), I am computing a DualMat with parmetis, saving it into a file. Then I recompile the code (using a petsc compiled with ptscotch), an load the DualMat from a file rather then forming a new one. I did a successful test when running on one processor. but I am having trouble when try on more.
I though the the dual graph was computed only once, even during the mpi process, instead it seems to be recomputed more than once. Not sure why.... sure i am missing something ???
--
You received this message because you are subscribed to the Google Groups "pflotran-dev" group.
To view this discussion on the web visit https://groups.google.com/d/msgid/pflotran-dev/CACGnotZfbdvEieXFn%2B6RYzj4o4yM_%3DK0NkHcFeK5CESL%2BovcKw%40mail.gmail.com.
Gautam Bisht
Hi Paolo,On Sun, Nov 3, 2013 at 3:53 AM, Paolo Orsini <paolo....@gmail.com> wrote:
Hi Jed,I found out what the problem is.First of all I printed all the matrices out, to be sure that MatConvert was working fine.And that was ok: AIJ_mat had the same connectivity table than Adj_mat.Then I compared the dual graph (stored in DualMat) computed by the Parmetis routine (MatMeshToCellGraph) and with MatMatTransposeMult. And they were quite different.The computation of the dual graph of the mesh is a bit more complicated than multiplying the adjacency matrix by its transpose, but not far off. With this operation, also the cells that share only one node are connected in the dual graph.Instead, the minimum number of common nodes is >1 (2 in 2D probelms, 3 in 3D problems). In fact, this is an input of MatMeshToCellGraph, I should have understood this before.This can be computed doing the transpose adjacency matrix (Adj_T), then doing the multiplication line by line of Adj time Adj_T, and discard the non zero entries coming from to elements that share a number of nodes less than the minimum number of common nodes imposed. I have not implement this yet, any suggestion is welcome.I also found out that Schotch has a facility to compute a dual graph from a mesh, but not PTScotch.Once the graph is computed, PTSchotch can load the central dual graph, and distribute it into several processors during the loading.Am i right to say that PETSC is interfaced only with PTSchotch and not with Scotch?To check if the PTSchotch partition works (within PFLOTRAN ), I am computing a DualMat with parmetis, saving it into a file. Then I recompile the code (using a petsc compiled with ptscotch), an load the DualMat from a file rather then forming a new one. I did a successful test when running on one processor. but I am having trouble when try on more.
I though the the dual graph was computed only once, even during the mpi process, instead it seems to be recomputed more than once. Not sure why.... sure i am missing something ???In PFLOTRAN, MatCreateMPIAdj() is called:- Once, if unstructured grid is specified in IMPLICIT format [in unstructured_grid.F90];
- Twice, if unstructured grid is specified in EXPLICIT format [in unstructured_grid.F90].
Paolo Orsini
Jed Brown
> Hi Jed,
>
> I am back to work on this now...
>
> Thanks for explaining how I can do this. it makes perfectly sense:
>
> A. Assign entries values = 1, when forming the Adj matrix,
>
> B. Convert the Adj matrix to AIJ matrix, so I can use MatMatTransposeMult
> (or form AIJ directly): can I preallocate AIJ before calling MatConvert? To
> avoid slow performance? Is there any conflict between the preallocoation
> functions and MatConvert?
> C. Filter AIJ, to remove entries with values <3 (for 3d problems). What
> function shall I use to do this operation?
another data structure? Anyway, I would just walk along calling
MatGetRow() and picking out the column indices that have values of at
least 3.
Paolo Orsini
Jed Brown
> MatConvert was slow when I tested 10 days ago, because was not
> preallocating the new matrix (see initial part of this thread). I was
> trying to find a work around.
> Unless you already fixed this, did you?... I haven't reinstalled petsc from
> then
function. What I recommended should work and be fast with 'next'. Can
you try that?
> I need the final Dual Graph (the filtered matrix) in Adj mat format.
> I will extract indices from the AIJ matrix (the matrix to filter), and I I
> will create a new Adj (avoiding to do an in-place operation as you
> suggested)
Jed Brown
> Thanks for the reminder, I forgot that we were missing a specialized
> function. What I recommended should work and be fast with 'next'. Can
> you try that?
Jed Brown
https://bitbucket.org/petsc/petsc/wiki/Home#markdown-header-following-petsc-development
Paolo Orsini
Jed Brown
> Hi Jed,
>
> I tried the MatConvert updated version, (from MPIAdj type to MPIAij type)
> but it seems still slow as before.
performance with a 3D Laplacian:
diff --git i/src/ksp/ksp/examples/tutorials/ex45.c w/src/ksp/ksp/examples/tutorials/ex45.c
index 4a67263..36f61d7 100644
--- i/src/ksp/ksp/examples/tutorials/ex45.c
+++ w/src/ksp/ksp/examples/tutorials/ex45.c
@@ -149,5 +149,13 @@ PetscErrorCode ComputeMatrix(KSP ksp,Mat jac,Mat B,MatStructure *stflg,void *ctx
ierr = MatAssemblyBegin(B,MAT_FINAL_ASSEMBLY);CHKERRQ(ierr);
ierr = MatAssemblyEnd(B,MAT_FINAL_ASSEMBLY);CHKERRQ(ierr);
*stflg = SAME_NONZERO_PATTERN;
+
+ {
+ Mat Badj,Baij;
+ ierr = MatConvert(B,MATMPIADJ,MAT_INITIAL_MATRIX,&Badj);CHKERRQ(ierr);
+ ierr = MatConvert(Badj,MATMPIAIJ,MAT_INITIAL_MATRIX,&Baij);CHKERRQ(ierr);
+ ierr = MatDestroy(&Badj);CHKERRQ(ierr);
+ ierr = MatDestroy(&Baij);CHKERRQ(ierr);
+ }
PetscFunctionReturn(0);
}
$ mpiexec -n 4 ./ex45 -da_grid_x 100 -da_grid_y 100 -da_grid_z 100 -ksp_max_it 1 -log_summary | great MatConvert
MatConvert 2 1.0 3.6934e-01 1.0 0.00e+00 0.0 1.6e+01 1.0e+04 1.9e+01 41 0 25 14 24 41 0 25 14 25 0
> Anyway, don't worry to much. I abandoned the idea of the conversion.
> I am forming the adjacency matrix directly in MPI AIJ format, so I don't
> have to do the conversion.
> I also realised that the Dual Graph is formed in parallel (in the current
> PFLOTRAN implementation), so I can't use the MatMatTransposeMult (which
> works only for sequential matrix).
>
> I will use instead MatCreatTranspose
just makes a matrix that behaves like the transpose.
> and MAtMatMult, then filtering the resulting matrix row by row and
> from the DualGraph. It will require more memory, but it should
> work. Do you see any problem with this?
Dan Zhou
Gautam Bisht
Dan Zhou
paolo....@gmail.com
Peter Lichtner
To view this discussion on the web visit https://groups.google.com/d/msgid/pflotran-dev/f83e4303-67df-4827-bf5d-ac8de192835a%40googlegroups.com.
Paolo Orsini
--
You received this message because you are subscribed to a topic in the Google Groups "pflotran-dev" group.
To view this discussion on the web visit https://groups.google.com/d/msgid/pflotran-dev/3D6736E5-A08E-447B-97DE-FE721625714A%40gmail.com.
paolo.t...@amphos21.com
El viernes, 1 de noviembre de 2013, 20:09:43 (UTC+1), Paolo Orsini escribió:
Hi Jed,I have tried to compile PFLOTRAN, after compiling petsc with ptscotch.The problem is that PFLOTRAN calls MatMeshToCellGraph to create the Dual graph, operation which is done by using the the parmetis function ParMETIS_V3_Mesh2Dual.Is there an equivalent function of MatMeshToCellGraph that uses PTScotch?Paolo
On Mon, Oct 7, 2013 at 9:03 PM, Jed Brown <jedb...@mcs.anl.gov> wrote:
Paolo Orsini
--
You received this message because you are subscribed to a topic in the Google Groups "pflotran-dev" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/pflotran-dev/MeGoaEvRHfs/unsubscribe.
To unsubscribe from this group and all its topics, send an email to pflotran-dev...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/pflotran-dev/c88a51d8-6898-422a-a6c2-1876ab92e738%40googlegroups.com.
Hammond, Glenn E
From: pflotr...@googlegroups.com <pflotr...@googlegroups.com>
On Behalf Of Paolo Orsini
Sent: Friday, October 25, 2019 6:12 AM
To: pflotran-dev <pflotr...@googlegroups.com>
Subject: [EXTERNAL] Re: [pflotran-dev: 5805] Re: [pflotran-users: 998] Distorted structured grid
Hi Paolo,
We build with PTScotch all the times, for the master branch, as ParMetis is open but not free.
However there are limitations, mainly you cannot use it with Implicit unstructured grids, as the domain decomposition of those type of meshes required a Parmetis function to create the dual graph (if I remember well).
Unless something has changed recently, as I haven't looked at the implicit grids recently.
However, for unstructured explicit mesh, which is the only ones we use, that function is not required, and you can build entirely without parmetis.
Decomposition of the “implicit” unstructured grids currently requires a call to MatMeshToCellGraph, which is wired to ParMETIS (https://www.mcs.anl.gov/petsc/petsc-current/docs/manualpages/MatOrderings/MatMeshToCellGraph.html). We would need to work with PETSc to have PT-SCOTCH replace it, if possible. I note that the PT-SCOTCH CeCILL-C license is compatible with GNU LGPL.
The “explicit” unstructured grid does not require ParMETIS as it calls MatPartitioningApply, which can leverage a number of partitioning types (https://www.mcs.anl.gov/petsc/petsc-current/docs/manualpages/Mat/MatPartitioningType.html#MatPartitioningType).
I would first look into gaining permission to use METIS/ParMETIS before looking into alternate partitioners.
Glenn
--
You received this message because you are subscribed to the Google Groups "pflotran-dev" group.
To unsubscribe from this group and stop receiving emails from it, send an email to
pflotran-dev...@googlegroups.com.
To view this discussion on the web visit
https://groups.google.com/d/msgid/pflotran-dev/CACGnotbtcgf8j%3D5pe8Hx0pLXTbjfy7b0vo5P5JRU0SOsfb30Aw%40mail.gmail.com.

