AW: [unmarked] Occu() issue. Hessian is singular/starting values
Marc Kery
Dear Isabelle,
here are a couple of quick ideas:
- It is often a good idea to build up a more complex model in steps of increasing complexity. That is, perhaps even start with the simplest model, with two intercepts for (in your case) psi and p, and then add one covariate at a time. And then use the solutions of the previous model in your progression as the inits for the current model
- In unmarked you must supply one initial value for each parameter in the model and in the same order in which they appear when you do “coef(fm)” (where fm is a fitted model object). You provide inits as a list, e.g., as follows: “inits <- c(-0.3333, 0.453)”. Then, when you fit the model, you add an argument “starts = inits” in the call to function occu.
- When you do this step-up approach of model building, you add in an NA for the new parameter that does not appear in the previous model in your progression. So, if the previous model is the Null model with the two intercepts for psi and p and your current model has one continuous covariate in the detection part of the model, then you would do something like “inits <- c(coef(fm)[1], NA, coef(fm)[2])”.
Best regards --- Marc
Von: unma...@googlegroups.com <unma...@googlegroups.com>
Im Auftrag von Isabelle Ranson
Gesendet: Montag, 6. Juni 2022 14:52
An: unmarked <unma...@googlegroups.com>
Betreff: [unmarked] Occu() issue. Hessian is singular/starting values
Hello,
I'm trying to use occu() to estimate the detection probability of trapping invasive crabs in 3 trap types over 5 areas. My data includes the calculated effort (catch per unit effort of total crabs caught/per trap soak night). The dataset contains over 4500 individual trap checks, and 35,000 invasive crabs removed, from 1 month of surveying. There are a lot of NAs.
The code is coming along nicely, but I got stuck trying to add my co variates. My best guess is that I need to add starting values, but I have no idea how to find them. The error message is requesting 9 starting values, I assumed those have to do with the 3 trapping methods, 5 areas, and effort. So I attempted to use my table to provide starting values. Is it asking for psi coefficients? I’m not sure how to go about finding those.
I’ve copied the code over and all warnings and errors, sorry if it’s a bit long. Most of these warnings are totally okay. Any advice on how to find starting values would be greatly appreciated! I’m new to unmarked.
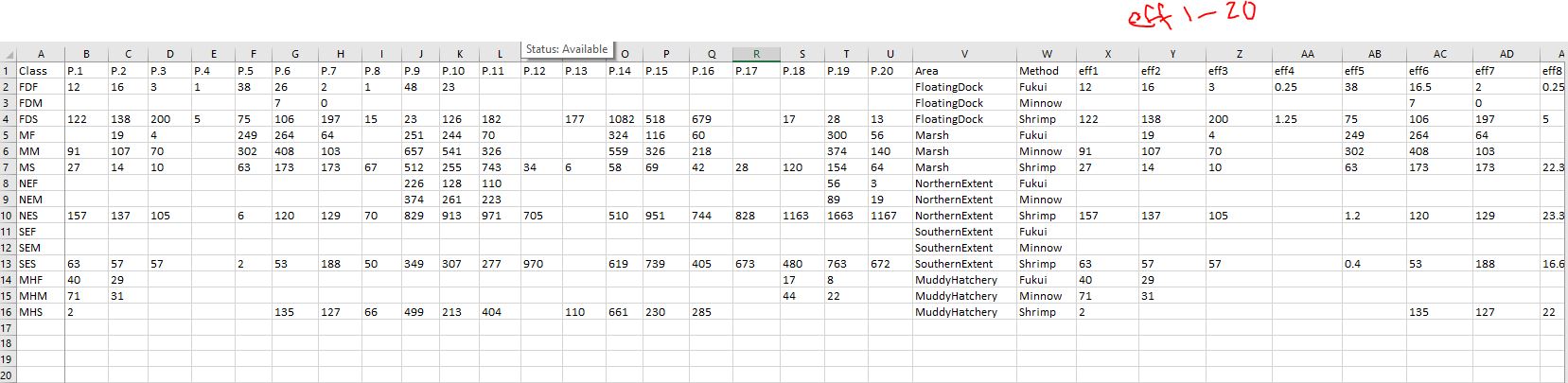
> egcdp <- isabelle_lisy[,c("P.1","P.2","P.3","P.4","P.5","P.6","P.7","P.8","P.9",
+ "P.10","P.11","P.12","P.13","P.14","P.15","P.16","P.17",
+ "P.18","P.19","P.20")]
> egcdp.umf <- unmarkedFrameOccu(y = egcdp,
+ siteCovs = data.frame(Method = isabelle_lisy$Method,
+ Area = isabelle_lisy$Area),
+ obsCovs = list(effort = isabelle_lisy[,c("eff1",
+ "eff2","eff3","eff4","eff5","eff6","eff7","eff8",
+ "eff9","eff10","eff11","eff12","eff13","eff14",
+ "eff15","eff16","eff17","eff18","eff19","eff20"),]))
> #this second model adds all co variates
> EGC2 <- occu(~Method +Area +effort ~1,egcdp.umf)
Warning messages:
1: 2 sites have been discarded because of missing data.
2: In truncateToBinary(designMats$y) :
Some observations were > 1. These were truncated to 1.
3: Hessian is singular. Try providing starting values or using fewer covariates.
> EGC2 <- occu(~Method +Area +effort ~1,egcdp.umf, starts= c())
Error in occu(~Method + Area + effort ~ 1, egcdp.umf, starts = c()) :
The number of starting values should be 9
In addition: Warning messages:
1: 2 sites have been discarded because of missing data.
2: In truncateToBinary(designMats$y) :
Some observations were > 1. These were truncated to 1.
#9 starting values? one for each of the 3 methods, 5 areas, and effort?
#grabbed the first value for each in the table
> EGC2 <- occu(~Method +Area +effort ~1,egcdp.umf, starts= c(12, 7, 122, 12, 19, 40, 226, 63, 12))
Warning messages:
1: 2 sites have been discarded because of missing data.
2: In truncateToBinary(designMats$y) :
Some observations were > 1. These were truncated to 1.
3: Hessian is singular. Try providing starting values or using fewer covariates.
#maybe it wanted the names instead?
> EGC2 <- occu(~Method +Area +effort ~1,egcdp.umf, starts= c("Fukui","Minnow",
+ "Shrimp","FloatingDock","Marsh","MuddyHatchery","NorthernExtent",
+ "SouthernExtent","effort"))
Error in optim(starts, nll, method = method, hessian = se, ...) :
non-finite value supplied by optim
In addition: Warning messages:
1: 2 sites have been discarded because of missing data.
2: In truncateToBinary(designMats$y) :
Some observations were > 1. These were truncated to 1.
3: In optim(starts, nll, method = method, hessian = se, ...) :
NAs introduced by coercion


--
*** Three hierarchical modeling email lists ***
(1) unmarked (this list): for questions specific to the R package unmarked
(2) SCR: for design and Bayesian or non-bayesian analysis of spatial capture-recapture
(3) HMecology: for everything else, especially material covered in the books by Royle & Dorazio (2008), Kéry & Schaub (2012), Kéry & Royle (2016, 2021) and Schaub & Kéry (2022)
---
You received this message because you are subscribed to the Google Groups "unmarked" group.
To unsubscribe from this group and stop receiving emails from it, send an email to
unmarked+u...@googlegroups.com.
To view this discussion on the web visit
https://groups.google.com/d/msgid/unmarked/7cc84a4a-e5d6-4113-b917-1a106240914an%40googlegroups.com.
Jim Baldwin
results <- occu(~Method ~1, egcdp.umf)# Call:# occu(formula = ~Method ~ 1, data = ufo)## Occupancy:# Estimate SE z P(>|z|)# 15.4 598 0.0257 0.98## Detection:# Estimate SE z P(>|z|)# (Intercept) 11.81 65 0.181732 0.856# MethodMinnow -8.64 65 -0.132830 0.894# MethodShrimp 12.65 15006 0.000843 0.999## AIC: 16.39769
# psi1-1/(1+exp(results@opt$par[1]))# 0.9999998# p(Fukui)1-1/(1+exp(results@opt$par[2]))# 0.9999926# p(Minnow)1-1/(1+exp(results@opt$par[2]+results@opt$par[3]))# 0.9600037# p(Shrimp)1-1/(1+exp(results@opt$par[2]+results@opt$par[4]))# 1
To view this discussion on the web visit https://groups.google.com/d/msgid/unmarked/ZR0P278MB08692CF3C217EF4B2E2D50BFEBA59%40ZR0P278MB0869.CHEP278.PROD.OUTLOOK.COM.
Isabelle Ranson
"month.7","month.8","month.9","month.10","month.11",
"month.12","month.13","month.14","month.15","month.16",
"month.17","month.18","month.19","month.20")]
siteCovs = NULL,
obsCovs = list(Method = EGC_sept[,c("method.1",
"method.2","method.3","method.4","method.5","method.6",
"method.7","method.8","method.9","method.10","method.11",
"method.12","method.13", "method.14","method.15",
"method.16","method.17","method.18","method.19",
"method.20")],
region = EGC_sept[,c("area.1","area.2","area.3",
"area.4","area.5","area.6","area.7","area.8","area.9",
"area.10","area.11","area.12", "area.13","area.14",
"area.15","area.16","area.17","area.18", "area.19",
"area.20")],
effort = EGC_sept[,c("effort.1","effort.2",
"effort.3","effort.4","effort.5","effort.6","effort.7",
"effort.8","effort.9","effort.10","effort.11",
"effort.12","effort.13","effort.14","effort.15",
"effort.16","effort.17","effort.18","effort.19",
"effort.20"),]),
type ="removal")
