unmarked single season occupancy fits well, but I get errors with mb.gof.test
96 views
Skip to first unread message
Airy González
Oct 5, 2023, 12:23:25 AM10/5/23
to unmarked
I fit single-season occupancy models for a Leporidae dataset where I have 115 sites from which 25 were double surveyed. After modSel() I got the best model was 'psi(elevation + yearBefore_coldestQTmean) p(forbCover + wind + (1|observer)).'
I perform VIF, given I know that temperature increases with elevation, and I wonder whether there might be multicollinearity. VIF came back at 4.3; low. There are no errors or warnings up to here.
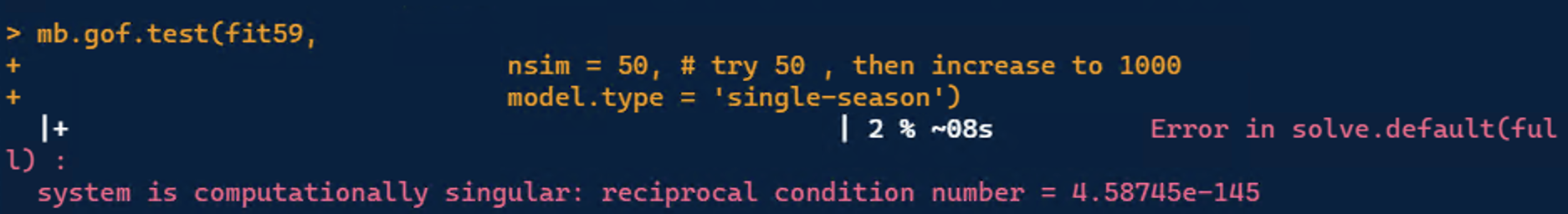
Next, I proceeded to perform GOF, and I am getting this error. Does anyone know the reason why the simulations are not running? What would the next step would be to debug this? Perhaps I needed a lower value than 4.3 for my VIF, and maybe I should regress yearBefore_coldestQTmean on elevation?
Thanks for your time,
~Airy![]()
![]()

Jim Baldwin
Oct 5, 2023, 1:06:55 AM10/5/23
to unma...@googlegroups.com
"Bad" condition numbers are usually associated with trying to take inverses of nearly singular matrices. So I would check the parameter correlation matrix for entries that are very close to +1 or -1 as an initial diagnostic. If none of the off-diagonal correlations are near +1 or -1, then there is most likely some other issue causing the trouble.
There is probably some very nice function in umarked to obtain the parameter correlation matrix but here is a brute force approach (assuming the the occu results are in fm):
covmat <- fm@opt$hessian
cormat <- covmat
for (i in 1:dim(covmat)[1]) {
for (j in i:dim(covmat)[1]) {
if (i==j) {
cormat[i, i] <- 1
} else {
cormat[i, j] <- covmat[i, j]/sqrt(covmat[i,i]*covmat[j,j])
cormat[j, i] <- cormat[i, j]
}
}}
cormat
cormat <- covmat
for (i in 1:dim(covmat)[1]) {
for (j in i:dim(covmat)[1]) {
if (i==j) {
cormat[i, i] <- 1
} else {
cormat[i, j] <- covmat[i, j]/sqrt(covmat[i,i]*covmat[j,j])
cormat[j, i] <- cormat[i, j]
}
}}
cormat
Jim
--
*** Three hierarchical modeling email lists ***
(1) unmarked (this list): for questions specific to the R package unmarked
(2) SCR: for design and Bayesian or non-bayesian analysis of spatial capture-recapture
(3) HMecology: for everything else, especially material covered in the books by Royle & Dorazio (2008), Kéry & Schaub (2012), Kéry & Royle (2016, 2021) and Schaub & Kéry (2022)
---
You received this message because you are subscribed to the Google Groups "unmarked" group.
To unsubscribe from this group and stop receiving emails from it, send an email to unmarked+u...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/unmarked/51c210eb-a261-4264-a071-e841cf757e52n%40googlegroups.com.
Jim Baldwin
Oct 5, 2023, 7:07:49 PM10/5/23
to unma...@googlegroups.com
Sorry, I left off the "solve" function in the first line of the code. Here's the correct code to obtain the correlation matrix for all estimated parameters:
covmat <- solve(fm@opt$hessian)
cormat <- covmatfor (i in 1:dim(covmat)[1]) {for (j in i:dim(covmat)[1]) {if (i==j) {cormat[i, i] <- 1} else {cormat[i, j] <- covmat[i, j]/sqrt(covmat[i,i]*covmat[j,j])cormat[j, i] <- cormat[i, j]}}}cormat
The correlation matrix for just the occupancy parameters and just the detection parameters can be obtained more directly with
fm@estimates@estimates$state@covMatfm@estimates@estimates$det@covMat
But if there's an issue that might be explained with large correlations, then I would go with examining all of the correlations of all estimated parameters.
Jim
Airy González
Oct 6, 2023, 12:14:29 AM10/6/23
to unma...@googlegroups.com
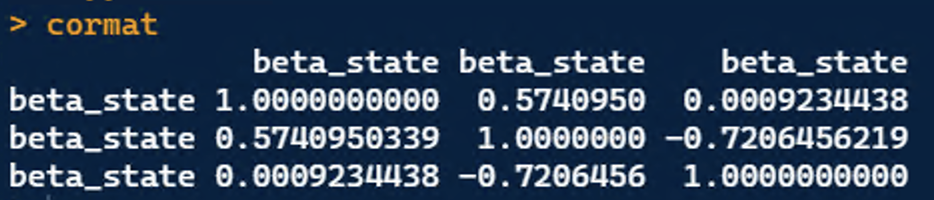
Jim, that helps a lot. I couldn't access the 'fm@opt$hessian, but I was able to access the correlation matrices for 'state' and 'det.' I don't have values near +1 or -1 on the off-diagonal correlations (See the cormat for 'state' below; the cormat from 'det' is too big to share). If multicollinearity is not the problem. What else could be?
Thanks for your help,
~Airy
To view this discussion on the web visit https://groups.google.com/d/msgid/unmarked/CAJCKiww5LwyaX3aJihZt%2BAXsMceV_3UBgjxZhO%2BH%3DtG%3DE3DQKQ%40mail.gmail.com.
Airy Adriana Peralta
Ph. D. Student & Professional Aide
Ecology and Evolutionary Biology
Institute of Arctic and Alpine Research - CU Boulder
Reply all
Reply to author
Forward
0 new messages
