Code related to ADVI algorithm
Visto 108 veces
Saltar al primer mensaje no leído
lala z
8 may 2022, 21:41:318/5/22
a TVB Users
Hello, is there any code related to advi algorithm in bvep library? I didn't find it
WOODMAN Michael
9 may 2022, 3:45:049/5/22
a tvb-...@googlegroups.com
hi
ADVI is a variation inference algorithm implemented by Stan. PyMC3 and other toolboxes demo'd in the BVEP repository also implement variational inference. In other words, it is the toolbox, but not the modeler who implements ADVI, and all the
models in BVEP should already be usable with ADVI.
cheers,
Marmaduke
On 9 May 2022, at 03:41, 'lala z' via TVB Users <tvb-...@googlegroups.com> wrote:
Hello, is there any code related to advi algorithm in bvep library? I didn't find it
--
You received this message because you are subscribed to the Google Groups "TVB Users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to tvb-users+...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/tvb-users/bb669850-338d-4720-856c-c85c3ea36d76n%40googlegroups.com.
lala z
12 may 2022, 9:35:0212/5/22
a TVB Users
OK, thank you very much for your reply.
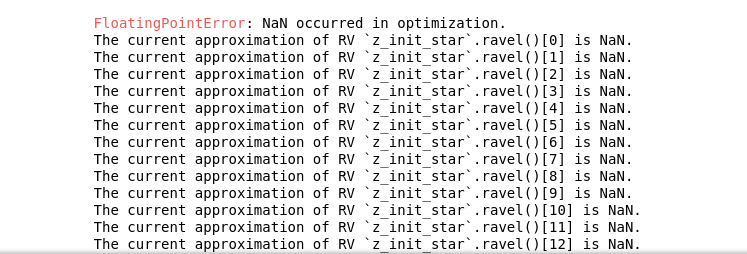
I'm using ADVI algorithm with pymc3 in BVEP model recently, but I found a new problem when I use non-central
parametrization and give a distribution to the parameters eps and sig, As shown below:

the following error will appear:

But when I give eps and sig a certain value, for example, eps = 0.06, sig = 0.16, there will be no such problem, and there will be no such problem when using central parameterization?As you said, would it be better to use central parameterization for ADVI?
WOODMAN Michael
12 may 2022, 10:16:5512/5/22
a tvb-...@googlegroups.com
Hi,
I'd guess this is happening because sig gets too large, destabilizing the time stepping in the non-centered case. This doesn't occur if you fix the value because it cannot change, and it doesn't happen in the centered parameterization because
the data directly inform (thus constrain) each time step independently. You could try to print(sig) in the model to see if that's what's happening.
General advice is to use non-centered parameterizations because they reduce nonlinear correlations between parameters that the sampler cannot handle well, but this advice may not apply to all models with ADVI or other algorithms.
cheers,
Marmaduke
On 12 May 2022, at 15:35, 'lala z' via TVB Users <tvb-...@googlegroups.com> wrote:
OK, thank you very much for your reply.I'm using ADVI algorithm with pymc3 in BVEP model recently, but I found a new problem when I use non-central parametrization and give a distribution to the parameters eps and sig, As shown below:
<7273c6e6df6d77a9870e974706113a6.png>
the following error will appear:
<06ccb1a27b367d18e75a2dafd461caa.png>But when I give eps and sig a certain value, for example, eps = 0.06, sig = 0.16, there will be no such problem, and there will be no such problem when using central parameterization?As you said, would it be better to use central parameterization for ADVI?
在2022年5月9日星期一 UTC+8 15:45:04<marmaduke.woodman> 写道:
hi
ADVI is a variation inference algorithm implemented by Stan. PyMC3 and other toolboxes demo'd in the BVEP repository also implement variational inference. In other words, it is the toolbox, but not the modeler who implements ADVI, and all the models in BVEP should already be usable with ADVI.
cheers,Marmaduke
On 9 May 2022, at 03:41, 'lala z' via TVB Users <tvb-...@googlegroups.com> wrote:
Hello, is there any code related to advi algorithm in bvep library? I didn't find it
--
You received this message because you are subscribed to the Google Groups "TVB Users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to tvb-users+...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/tvb-users/bb669850-338d-4720-856c-c85c3ea36d76n%40googlegroups.com.
--
You received this message because you are subscribed to the Google Groups "TVB Users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to tvb-users+...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/tvb-users/c67fa74d-b759-4f0f-9d75-dfae0489ac6dn%40googlegroups.com.
<06ccb1a27b367d18e75a2dafd461caa.png><7273c6e6df6d77a9870e974706113a6.png>
lala z
13 may 2022, 2:29:3813/5/22
a TVB Users
Hello, I tried to use central parameterization in advi algorithm, but the final result is very poor. What you said is to print the value of sig after sampling with advi algorithm, and then sample the value of sig from a posteriori distribution to see if the value of sig is too large?
meysam....@gmail.com
13 may 2022, 5:22:3913/5/22
a TVB Users
hi,
To print the value of parameters for debugging, you can use
import theano.tensor as tt
sig_print = tt.printing.Print("sig")(sig)
see an example here:
However, if you fixed them it would be easier and faster to get convergence. You can drop them from parameters and just assign fixed values.
lala z
15 may 2022, 21:40:3915/5/22
a TVB Users
Hello, this is the result of using the advi algorithm in pymc3, but I feel that the results of posterior z-scores is not very good. Have you tried the advi algorithm in pymc3, and this situation has also occurred? Maybe there is any way to improve it?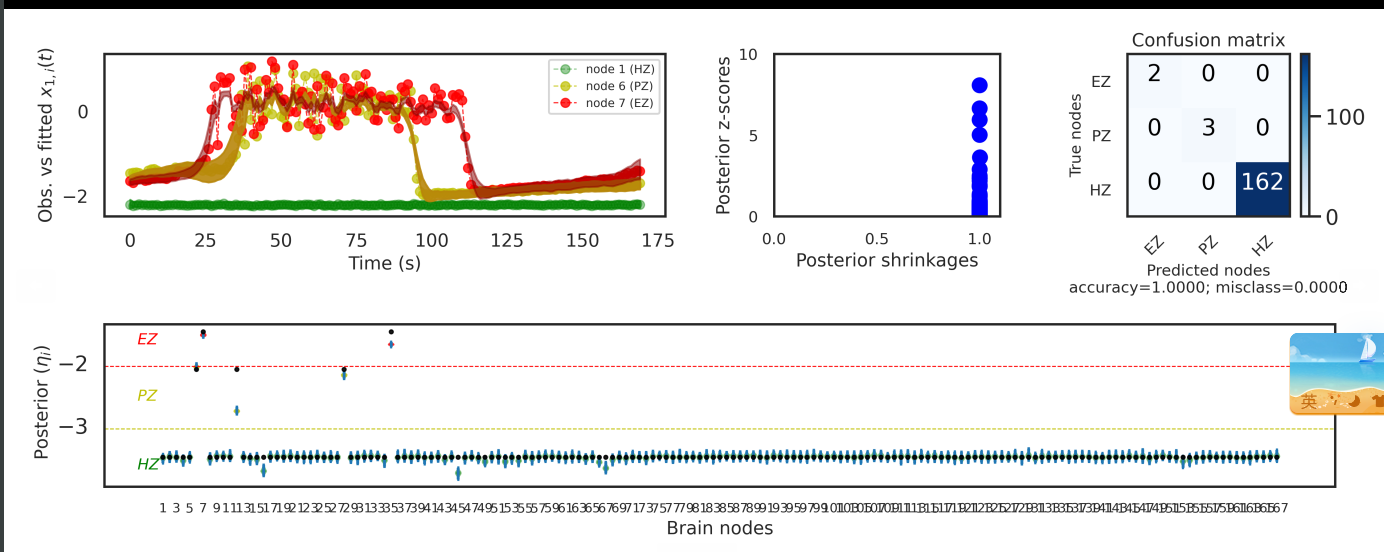

lala z
18 may 2022, 4:46:2718/5/22
a TVB Users
Hello, I see that you have added the code for VEP in Pyro in the pyro-vi branch on the BVEP repository . What function does this code realize? Can it realize the estimation of relevant parameters like the model in BVEP, but I don't seem to find that the epileptogenic index η is defined in it,
WOODMAN Michael
20 may 2022, 3:39:1520/5/22
a tvb-...@googlegroups.com
Hi,
This Pyro code, primarily a proof of concept for full-cohort variational inference, implements an autoregressive model for source activity, not the VEP model per se, though it could be added, in the model component here,
Once the source estimates are obtained, one would be able to compute some index on them to choose which are epileptogenic.
cheers,
Marmaduke
To view this discussion on the web visit https://groups.google.com/d/msgid/tvb-users/c180111c-4c82-43ce-a91f-b49682112809n%40googlegroups.com.
lala z
30 may 2022, 8:56:2030/5/22
a TVB Users
Hello, I'm trying to build the BVEP model with Pyro and use Bayesian inference. I use SVI in Pyro to approximate the posterior distribution. I use the central parameterization because the result of non-central parameterization seems very bad, but there is also a strange problem. The result is as follows: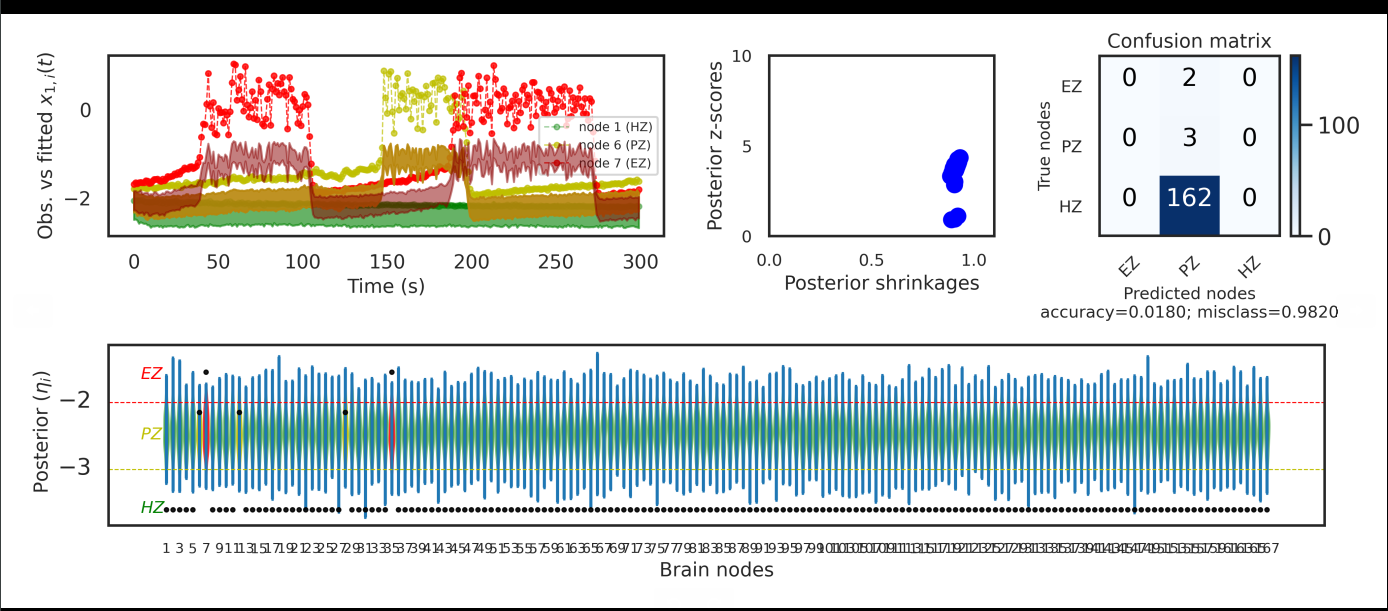
As shown in the figure, the predicted time series and the observed values seem to have the same shape, but the predicted time series' values are generally lower than the values of observed , and the results of the confusion matrix are also very poor. I guess it may be caused by the inconsistency between the predicted time series and the observed values. This problem has troubled me for a long time. I really don't know why. Is BVEP not suitable for using Pyro? I would appreciate it if you could give me some advice.

As shown in the figure, the predicted time series and the observed values seem to have the same shape, but the predicted time series' values are generally lower than the values of observed , and the results of the confusion matrix are also very poor. I guess it may be caused by the inconsistency between the predicted time series and the observed values. This problem has troubled me for a long time. I really don't know why. Is BVEP not suitable for using Pyro? I would appreciate it if you could give me some advice.
WOODMAN Michael
30 may 2022, 9:10:1530/5/22
a tvb-...@googlegroups.com
Hi,
It looks like the model is not implemented correctly, as the posterior is not conditioned at all with respect to the x values. Can you share your code implementing BVEP in Pyro? .
cheers,
Marmaduke
On 30 May 2022, at 14:56, 'lala z' via TVB Users <tvb-...@googlegroups.com> wrote:
Hello, I'm trying to build the BVEP model with Pyro and use Bayesian inference. I use SVI in Pyro to approximate the posterior distribution. I use the central parameterization because the result of non-central parameterization seems very bad, but there is also a strange problem. The result is as follows:
To view this discussion on the web visit https://groups.google.com/d/msgid/tvb-users/d750b49d-9740-4241-a833-70f941766cden%40googlegroups.com.
<46681e6bebf00d81c4eb0de0457e668.png>
lala z
30 may 2022, 9:15:2630/5/22
a TVB Users
Thank you for your reply. I will send my code to your email
Se ha eliminado el mensaje
meysam....@gmail.com
30 may 2022, 10:12:2230/5/22
a TVB Users
Hi,
Your results are accurate according to confusion matrix.
If you wish to improve the posterior z-score, you can fixed eps, and sig, and for instance, here run it with larger eps fixed; The inference on these hyperparameters are challenging with ADVI/NUTS, though NUTS (in particular Stan implementation) with non-centered form of parameterization can handle better.
lala z
30 may 2022, 21:52:3630/5/22
a TVB Users
OK, thank you for your help. I will try your advice
Se ha eliminado el mensaje
lala z
1 jun 2022, 3:55:001/6/22
a TVB Users
Hello, I've been thinking for a long time. Maybe I still don't understand what you said that I don't need a for loop over time in the centered version if I use Pyro.I have
read carefully in the example code you provide .The sources in the example code initially sampled data with the size of (n_node, n_time) from the uniform distribution. However, for the VEP model, if the for loop is not used, how can I generate the simulated time series data to perform Bayesian inversion through the observed data?
The BVEP central parameterization model I built with Pyro refers to the central parameterization model in pymc3 provided by you. Maybe I still lack some knowledge in this field. I don't know the difference between building a central parameterization model in Pyro and pymc3. I hope you can give me some suggestions.
Finally, I would like to ask if I want to use more seizure data to infer the epileptogenic area, whether the SVI in pyro will be faster than the ADVI in pymc3, but at present, it seems that the ADVI in pymc3 is faster.
Thank you for your patient reply during this time
Thank you for your patient reply during this time
在2022年5月30日星期一 UTC+8 22:12:22<meysam....@gmail.com> 写道:
WOODMAN Michael
1 jun 2022, 4:27:511/6/22
a 'lala z' via TVB Users
hi,
Your model code has to evaluate the likelihood of the parameters given your data. In the case of the centered parametrization, the neural time series are a subset of your parameters, and then you just need to evaluate the likelihood of the time
series given your observations, and the likelihood of the transition between neural states, which for an SDE is a normal distribution where the mean is given by the deterministic part of the SDE, and the standard deviation is from the drift part of the SDE.
This can be evaluated in a vectorized fashion (i.e. every time point at the same time), which is especially helpful for long time series or large datasets, since parallelization on GPUs can be used.
In contrast, the non-centered form has the noise process as a subset of the parameters, and then during likelihood evaluation, one integrates the SDE in a normal fashion. The rest is the same.
On performance, it will depend on your data mainly. If ADVI in PyMC3 is working for you, then you should use that.
cheers,
Marmaduke
lala z
1 jun 2022, 8:24:021/6/22
a TVB Users
Hello, my understanding is that in central parameterization, the value of a point in the time series depends on the value of the previous time point. For example, the value of X (t) is sampled from the normal distribution (mean, std), and mean=step_ ode(x (t-1)), std is equal to the drift part of SDE. So set an initial value for each brain node, and then use step_ ode () can generate the estimated value of a time series within the time that consistent with the observed data, and then estimate the parameters through the observed data. I wonder if my understanding here is correct.
What I really don't understand is what the vectorized fashion is, that is, how the vectorized fashion realizes the above process.Does every time point at the same time mean that the dependency of each value in the time series is ignored?
I would appreciate it if you could help me solve this problem that has puzzled me for a long time!
What I really don't understand is what the vectorized fashion is, that is, how the vectorized fashion realizes the above process.Does every time point at the same time mean that the dependency of each value in the time series is ignored?
I would appreciate it if you could help me solve this problem that has puzzled me for a long time!
WOODMAN Michael
2 jun 2022, 11:21:102/6/22
a TVB Users
Hi,
The time dependency is not ignored. I suspect your confusion arises because you are thinking in terms of simulation: a loop in time is required for simulation because the next time steps is not available until is it computed from the previous
time step.
When you estimating fitting an SDE, in the centered parametrization, the states at each time point are part of the parameter space. Let's take an AR(1) process like this
x_{t+1} = a * x_t + e(t)
In forward simulation (and the non-centered parametrization), you have 1 parameter (namely, a) and you choose x_{t=0} and sample a normal noise process e(t) to step through time.
In the non-centered parameterization, for 10 time points, you have 11 parameters, namely a, x_{t=0} and 9 parameters for the realized values of e(t). The code then computes x_{t+1} stepping through time iteratively.
In the centered parametrization, for 10 time points, you have 11 parameters, namely a, and 10 parameters for the realized values of x_{t}. The code does not compute x_{t+1} iteratively because they are already available as part of the parameters,
and instead the code computes how well the values of x_{t} match the AR(1) process described above, in terms of a normal distribution,
x_{t+1} ~ N(a * x_t, 1)
This is where the dependence in time enters explicitly, but because all values of x_t are available, this can be done in a vectorized fashion without the for loop in time.
The two different parametrizations induce different correlations between parameters: in the non-centered case of 9 parameters from an i.i.d. normal, the parameters are supposedly uncorrelated and easy to estimate. In the centered case of 10 parameters,
they could be highly nonlinearly correlated, which can make estimate difficult or impossible. This is why the non-centered case should, theoretically, provide better estimates, but in practice, both can be tested.
cheers,
Marmaduke
WOODMAN Michael
2 jun 2022, 11:27:592/6/22
a TVB Users
hi
I should have added that the line below
x_{t+1} ~ N(a * x_t, 1)
is not an assignment in code, it's a probability evaluation. In Python, it would be equivalent to something like
logp += normal.lpdf(x[1:], a * x[:-1], 1)
where logp is the objective function to maximize (for optimization).
cheers,
Marmaduke
lala z
6 jun 2022, 3:54:286/6/22
a TVB Users
Hello, thank you very much for your patient reply, which is very helpful to me, but this concept is still a little abstract to me. so I tried to change the code and use the vectorization method. I don't know if this is correct.The result is still bad after this modification .Can you give me some suggestions?


WOODMAN Michael
7 jun 2022, 3:42:287/6/22
a TVB Users
hi
I don't think plate is required, and your code still doesn't make use of the ODE, so you need something more like
# sample x w/ uninformative prior
z = torch.zeros((nt, 2, nn))
x_dist = Normal(z, z+3).independent(3)
x = pyro.sample('x', x_dist)
# compute transitions according to step_ode (all time points)
nx = step_ode(x, ...)
# condition x on transitions
step_dist = Normal(nx[:-1], sqrt(dt)*sig).independent(3)
step_samp = pyro.sample('step_samp', step_dist,
obs=x[1:])
which is what's done in the sample code.
cheers,
Marmaduke
On 6 Jun 2022, at 09:54, 'lala z' via TVB Users <tvb-...@googlegroups.com> wrote:
Hello, thank you very much for your patient reply, which is very helpful to me, but this concept is still a little abstract to me. so I tried to change the code and use the vectorization method. I don't know if this is correct.The result is still bad after this modification .Can you give me some suggestions?
<310221577cc8b869bb096d2efe4b6ec.png>
在2022年6月2日星期四 UTC+8 23:27:59<marmaduke.woodman> 写道:
hi
I should have added that the line below
x_{t+1} ~ N(a * x_t, 1)
is not an assignment in code, it's a probability evaluation. In Python, it would be equivalent to something like
logp += normal.lpdf(x[1:], a * x[:-1], 1)
where logp is the objective function to maximize (for optimization).
cheers,Marmaduke
On 2 Jun 2022, at 17:21, WOODMAN Michael <marmaduk...@univ-amu.fr> wrote:
instead the code computes how well the values of x_{t} match the AR(1) process described above, in terms of a normal distribution,
x_{t+1} ~ N(a * x_t, 1)
This is where the dependence in time enters explicitly, but because all values of x_t are available, this can be done in a vectorized fashion without the for loop in time.
--
You received this message because you are subscribed to the Google Groups "TVB Users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to tvb-users+...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/tvb-users/222ee41b-1444-434a-b015-72a7104d061dn%40googlegroups.com.
<310221577cc8b869bb096d2efe4b6ec.png>
Se ha eliminado el mensaje
lala z
7 jun 2022, 8:29:387/6/22
a TVB Users
Thank you very much for your help. I finally figured out this method. It's very clever. I admire your wisdom!
WOODMAN Michael
7 jun 2022, 8:32:297/6/22
a TVB Users
hi
Glad to read that it makes sense. We got the technique from the Stan User Guide, which can be a helpful read when starting out on Bayesian inference.
cheers,
Marmaduke
lala z
7 jun 2022, 8:42:127/6/22
a TVB Users
OK, I'll read it later.
Thank you,
lala
lala z
8 jun 2022, 23:01:438/6/22
a TVB Users
Hello
Second, compute transitions according to step_ode():


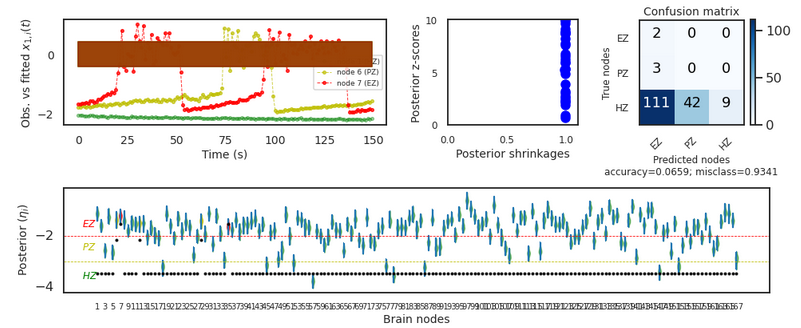
A bad news. After I modified the code according to your suggestion, the result was still very bad,I don't know if there is something wrong with my code. The code I modified is as follows:
First, x and z are uninformative priors:

Second, compute transitions according to step_ode():

Third,condition x and z on transitions

The code for step_ode() is as follows:

The results are shown in the following figure

Is there something wrong with my code?
Thank you,
lala
WOODMAN Michael
9 jun 2022, 3:30:359/6/22
a TVB Users
hi,
yes, the difference coupling is wrong, it should read
diff_state = from_state - to_state
as the summand is x_j - x_i with a sum over j.
see the Stan code or others for reference,
cheers,
Marmaduke
lala z
9 jun 2022, 9:03:119/6/22
a TVB Users
Hello,

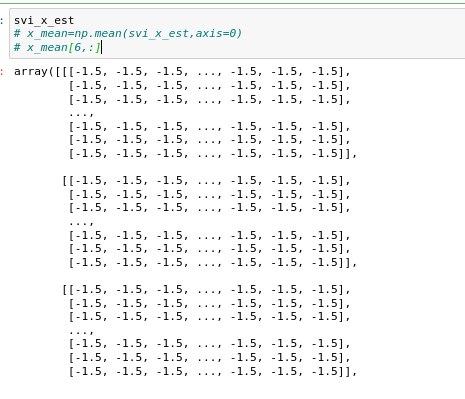
I have corrected it, but the result is still not ideal. At first, I set both x and z as uninformational priors, as shown below:

Then I printed out that all values of X sampled from the posterior distribution are 0.
When the priors of X and Z are sampled from Normal (-1.5 , 0) and Normal (-3.5 , 0) as follows:

all values of X sampled from the posterior distribution are -1.5, as follows:

I feel like step_ ode() doesn't work, Do you have any ideas?
Thank you ,
lala
WOODMAN Michael
9 jun 2022, 11:05:589/6/22
a TVB Users
hi,
I agree that there's a bug, but I don't see your whole code, so you'll have to keep looking. I would suggest sticking to the validated codes that Meysam provided under PyMC3 or Stan.
cheers,
Marmaduke
On 9 Jun 2022, at 15:03, 'lala z' via TVB Users <tvb-...@googlegroups.com> wrote:
Hello,I have corrected it, but the result is still not ideal. At first, I set both x and z as uninformational priors, as shown below:
<881e3c689efeb2845eb087397ab372b.png>Then I printed out that all values of X sampled from the posterior distribution are 0.When the priors of X and Z are sampled from Normal (-1.5 , 0) and Normal (-3.5 , 0) as follows:
<fad9bcab5dec1731aa832c21cc4e948.png>
all values of X sampled from the posterior distribution are -1.5, as follows:
<b144634ba05bf86a2771126c9dd09f8.png>
I feel like step_ ode() doesn't work, Do you have any ideas?
Thank you ,lala在2022年6月9日星期四 UTC+8 15:30:35<marmaduke.woodman> 写道:
hi,
yes, the difference coupling is wrong, it should read
diff_state = from_state - to_state
as the summand is x_j - x_i with a sum over j.
see the Stan code or others for reference,
cheers,Marmaduke
On 9 Jun 2022, at 05:01, 'lala z' via TVB Users <tvb-...@googlegroups.com> wrote:
Is there something wrong with my code?
--
You received this message because you are subscribed to the Google Groups "TVB Users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to tvb-users+...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/tvb-users/bd800fb3-00dc-4463-9e78-bb071f922a1fn%40googlegroups.com.
<881e3c689efeb2845eb087397ab372b.png><fad9bcab5dec1731aa832c21cc4e948.png><b144634ba05bf86a2771126c9dd09f8.png>
lala z
10 jun 2022, 2:09:1910/6/22
a TVB Users
Hello, I have checked my code repeatedly, and it should be correct. I also don't know why this problem occurs. Maybe I should write a manual guide instead of using an automatic guide?Is BVEP not applicable to Pyro?
WOODMAN Michael
10 jun 2022, 3:15:4510/6/22
a TVB Users
hi,
> On 10 Jun 2022, at 08:09, 'lala z' via TVB Users <tvb-...@googlegroups.com> wrote:
>
> Hello, I have checked my code repeatedly, and it should be correct. I also don't know why this problem occurs.
with all due respect, this happens to everyone; some bugs are harder than others to identify.
if you share the full source, perhaps privately if you prefer, I can look again.
> Maybe I should write a manual guide instead of using an automatic guide?
a guide is Pyro's word for variational distribution. You are already using the standard mean field normal variational distribution, which is used in PyMC3 and Stan, among others. A custom guide would be helpful if you know a priori specific things required such as Gaussian mixture.
> Is BVEP not applicable to Pyro?
It's not an issue of BVEP or Pyro, in my opinion, but a bug in the model code.
cheers,
Marmaduke
> On 10 Jun 2022, at 08:09, 'lala z' via TVB Users <tvb-...@googlegroups.com> wrote:
>
> Hello, I have checked my code repeatedly, and it should be correct. I also don't know why this problem occurs.
if you share the full source, perhaps privately if you prefer, I can look again.
> Maybe I should write a manual guide instead of using an automatic guide?
a guide is Pyro's word for variational distribution. You are already using the standard mean field normal variational distribution, which is used in PyMC3 and Stan, among others. A custom guide would be helpful if you know a priori specific things required such as Gaussian mixture.
> Is BVEP not applicable to Pyro?
cheers,
Marmaduke
lala z
10 jun 2022, 3:21:3810/6/22
a TVB Users
Hello,
Thank you very much for your reply. If you are willing to help me correct the problems in the code, I will be very grateful. I will share the whole code to your email.
Thanks,
lala
Responder a todos
Responder al autor
Reenviar
Se ha eliminado el mensaje
0 mensajes nuevos
