Cancer Imaging Archive Updates: February 2020
Tacconelli, Michelle (NIH/NCI) [C]
 |
||||||||||||||||||||||||||||||||||||||||
TCIA Update: February 2020 |
||||||||||||||||||||||||||||||||||||||||
|
The authors of "Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach" have recently shared a treasure trove of additional data from their study! In 2014 when the paper was published they shared the Lung3 cohort in TCIA under the name NSCLC-Radiomics-Genomics. Also in 2014, the Lung1 cohort was shared under the name NSCLC-Radiomics. RTSTRUCT contours of the gross tumor volumes were added in 2016. In Oct 2019 there were data corrections and DICOM-SEG objects added. Details can be seen on the "Versions" tab here. We are happy to announce the "Multiple delineation" cohort is now available as NSCLC-Radiomics-Interobserver1, including clinical data and the segmentations of primary tumors created by 5 different radiation oncologists (RTSTRUCT and SEG formats). The "H&N1" cohort is also now available as Head-Neck-Radiomics-HN1, including clinical data and radiation oncologist delineations of gross tumor volumes. Last but not least, they have released the "RIDER test/retest" data, which created segmentations from existing RIDER Lung CT "coffee break" studies on TCIA. |
||||||||||||||||||||||||||||||||||||||||
 |
||||||||||||||||||||||||||||||||||||||||
 |
||||||||||||||||||||||||||||||||||||||||
|
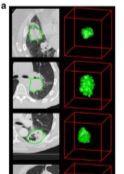
The new CT-ORG collection contains 140 patients' CT volumes with multiple organ segmentations (primarily lung, bones, liver, kidneys, bladder) in NifTI format to enable multi-class organ segmentation and competitive benchmarking of algorithms: https://doi.org/10.7937/tcia.2019.tt7f4v7o |
||||||||||||||||||||||||||||||||||||||||
 |
||||||||||||||||||||||||||||||||||||||||
 |
||||||||||||||||||||||||||||||||||||||||
|
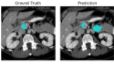
Excellent AI tutorial using the Pancreas-CT collection from TCIA. Learn more about this dataset at: https://dx.doi.org/10.7937/K9/TCIA.2016.tNB1kqBU |
||||||||||||||||||||||||||||||||||||||||
|
Here's an interesting project leveraging our API! The oncoramed "TCIA Bootstrap" repo on Github provides a small docker setup to quickly deploy and load TCIA collections into an OrthancServer DICOM server. Learn more at https://github.com/ oncoramedical/tcia_bootstrap |
||||||||||||||||||||||||||||||||||||||||
 |
||||||||||||||||||||||||||||||||||||||||
 |
||||||||||||||||||||||||||||||||||||||||
|
The ACRIN-NSCLC-FDG-PET clinical trial data previously under embargo is now open-access! Its objective was to assess SUV measurements from FDG-PET in lung cancer patients as a predictor of long-term clinical outcome after definitive chemoradiotherapy. https://doi.org/10.7937/tcia.2019.30ilqfcl |
||||||||||||||||||||||||||||||||||||||||
 |
The video recording & slides from last week's webinar about the proteogenomic analyses of the CPTAC-CCRCC (renal clear cell) cohort are available at: https:// wiki.cancerimagingarchive.net/display/Public/CPTAC+SIG+Webinars…. Hope you'll join us for the next one March 3 covering the LUAD (lung adenocarcinoma) cohort! |
|||||||||||||||||||||||||||||||||||||||
 |
Check out this tool for visualizing proteogenomic data from the CPTAC-CCRCC discovery project (renal clear cell): http://ccrcc.cptac-data-view.org. It also lets you easily view the related digitized histopathology slides on TCIA as you explore the heat map. |
|||||||||||||||||||||||||||||||||||||||
 |
||||||||||||||||||||||||||||||||||||||||
|
The QIN-Breast collection is now open-access! This dataset was originally released in 2015 and contains longitudinal PET/CT and quantitative MR images collected for the purpose of studying treatment assessment in breast cancer in the neoadjuvant setting: http://doi.org/10.7937/K9/TCIA.2016.21JUebH0 |
||||||||||||||||||||||||||||||||||||||||
 |
||||||||||||||||||||||||||||||||||||||||
|
2019 was a big year for the TCIA community! Submitters contributed to 37 datasets, bringing us to a total of 113. Site traffic surpassed 20,000 users/month. Downloads averaged 95 Tbytes/month, and 195 papers were published about TCIA datasets! https://cancerimagingarchive.net/publications/ |
||||||||||||||||||||||||||||||||||||||||
 |
||||||||||||||||||||||||||||||||||||||||
|
Working with our LIDC-IDRI collection? Instead of writing custom code to interpret the XML radiologist assessments, check out pylidc (https://pylidc.github.io) and the DICOM conversion of the annotation data (https://doi.org/10.7937/ TCIA.2018.h7umfurq) to get started more quickly. |
||||||||||||||||||||||||||||||||||||||||
|
Stay Connected with TCIA |
||||||||||||||||||||||||||||||||||||||||
