Adaptive treshold
125 views
Skip to first unread message
Mirko Sangiorgio
May 23, 2022, 5:04:57 AM5/23/22
to SpiNNaker Users Group
Hi, I'm trying to use SNN for classification using PYNN on SpiNNaker.
To create competition among neurons in the excitatory layer, I would like to use an adaptive treshold value ("v_tresh") instead of fixed value. I need that the every time a excitatory neuron spikes, the parameter "v_tresh" is increased for this specific neuron.
Is it possible?
Thanks.
Best regards, Mirko Sangiorgio.
Andrew Gait
May 23, 2022, 6:03:02 AM5/23/22
to Mirko Sangiorgio, SpiNNaker Users Group
Hi Mirko,
We don't have exactly what you're looking for in terms of moving the threshold value for a neuron when it spikes, but we do have an adaptive "additional input" model available which would I think give you the behaviour you're looking for with the right parameters.
You can use this adaptive model on a population as follows:
pop = sim.Population(n_neurons, sim.extra_models.IFCurrExpCa2Adaptive(**{"i_alpha": 0.1, "i_ca2": 0.0, "tau_ca2": 50.0}), label="pop")
The relevant parameters are shown with their default values - this was originally written with "calcium channels" in mind; i_ca2 is the "baseline current" from a channel, i_alpha is the amount of calcium "caused" by a spike, and tau_ca2 is used to determine
how to decay the adaptive input value back to its baseline value (via the formula exp(-t/tau_ca2)). In this instance, the more "calcium" added (i.e. the more positive you make i_alpha) leads to a
negative input value which means that the voltage value is reduced for a neuron after it spikes. I think this is what you would need to do to reduce the chances of a neuron spiking again in your excitatory layer - depending on your synapse weight values,
you may need to play with the parameter values in order to get the behaviour you are looking for. (Note that you can also make i_alpha negative which gives a
positive input value and increases the voltage for that neuron after spiking).
Any further questions about this, please let us know. (The adaptive "additional input" model is only available on LIF "current neurons" at the moment, but we can easily show you how to build a binary to use it with conductance or Izhikevich models if that's
what you're using instead).
Andy
----------------------------------------------------
Dr. Andrew Gait (he/him),
Dr. Andrew Gait (he/him),
Research Software Engineer,
APT Group,
Department of Computer Science,
The University of Manchester,
Oxford Road, Manchester M13 9PL
email: andre...@manchester.ac.uk
web: http://personalpages.manchester.ac.uk/staff/Andrew.Gait/
Department of Computer Science,
The University of Manchester,
Oxford Road, Manchester M13 9PL
email: andre...@manchester.ac.uk
web: http://personalpages.manchester.ac.uk/staff/Andrew.Gait/
I do not expect a reply to this email outside of normal working hours
From: spinnak...@googlegroups.com <spinnak...@googlegroups.com> on behalf of Mirko Sangiorgio <mirk...@hotmail.it>
Sent: 23 May 2022 09:54
To: SpiNNaker Users Group <spinnak...@googlegroups.com>
Subject: [SpiNNaker Mailing List] Adaptive treshold
Sent: 23 May 2022 09:54
To: SpiNNaker Users Group <spinnak...@googlegroups.com>
Subject: [SpiNNaker Mailing List] Adaptive treshold
--
You received this message because you are subscribed to the Google Groups "SpiNNaker Users Group" group.
To unsubscribe from this group and stop receiving emails from it, send an email to spinnakeruser...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/spinnakerusers/d86384c9-6247-4a78-aa4e-0bd919286401n%40googlegroups.com.
You received this message because you are subscribed to the Google Groups "SpiNNaker Users Group" group.
To unsubscribe from this group and stop receiving emails from it, send an email to spinnakeruser...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/spinnakerusers/d86384c9-6247-4a78-aa4e-0bd919286401n%40googlegroups.com.
mirkosan
May 23, 2022, 6:42:46 AM5/23/22
to SpiNNaker Users Group
Hi Andy, thank you for your reply.
I'm trying to replicate in a simple decoding task the work from Diehl PU, Cook M. Unsupervised learning of digit recognition using spike-timing-dependent plasticity. Front Comput Neurosci. 2015;9:99. Published 2015 Aug 3. doi:10.3389/fncom.2015.00099
Specifically I want to replicate what they call "Homoeostasis":
"The inhomogeneity of the input leads to different firing rates of
the excitatory neurons, and lateral inhibition further increases
this difference. However, it is desirable that all neurons have
approximately equal firing rates to prevent single neurons from
dominating the response pattern and to ensure that the receptive
fields of the neurons differentiate. To achieve this, we employ
an adaptive membrane threshold resembling intrinsic plasticity
(Zhang and Linden, 2003). Specifically, each excitatory neuron’s
membrane threshold is not only determined by v_thresh but by the
sum v_thresh + θ, where θ is increased every time the neuron fires
and is exponentially decaying (Querlioz et al., 2013). Therefore,
the more a neuron fires, the higher will be its membrane
threshold and in turn the neuron requires more input to spike
in the near future. Using this mechanism, the firing rate of
the neurons is limited because the conductance-based synapse
model limits the maximum membrane potential to the excitatory
reversal potential Eexc, i.e., once the neuron membrane threshold
is close to Eexc (or higher) it will fire less often (or even stop firing
completely) until θ decreases sufficiently "
I´m using
LIF "current neurons".
Did you understand better what I am looking for? Do you think that the adaptive model you suggested could be a good solution for my problem?
Thanks.
Mirko
Andrew Gait
May 23, 2022, 6:59:53 AM5/23/22
to mirkosan, SpiNNaker Users Group
Hi Mirko,
Yes, I think what I suggested could do effectively the same thing, but what it does is reduce the membrane voltage value rather than raise the threshold value (which amounts in effect to the same thing in terms of whether a neuron spikes or not). You just
need to work out what parameters they have used in the paper to determine θ and the decay rate of θ and substitute the relevant values in (as i_alpha and tau_ca where the decay is given by exp(-t/tau_ca).
Let us know if this works for you - if not then we'd like to know why, and we can then discuss other possible ways of solving your problem.
Andy
----------------------------------------------------
Dr. Andrew Gait (he/him),
Dr. Andrew Gait (he/him),
Research Software Engineer,
APT Group,
Department of Computer Science,
The University of Manchester,
Oxford Road, Manchester M13 9PL
email: andre...@manchester.ac.uk
web: http://personalpages.manchester.ac.uk/staff/Andrew.Gait/
Department of Computer Science,
The University of Manchester,
Oxford Road, Manchester M13 9PL
email: andre...@manchester.ac.uk
web: http://personalpages.manchester.ac.uk/staff/Andrew.Gait/
I do not expect a reply to this email outside of normal working hours
From: spinnak...@googlegroups.com <spinnak...@googlegroups.com> on behalf of mirkosan <mirk...@hotmail.it>
Sent: 23 May 2022 11:42
Sent: 23 May 2022 11:42
Subject: Re: [SpiNNaker Mailing List] Adaptive treshold
To view this discussion on the web, visit
https://groups.google.com/d/msgid/spinnakerusers/3fd98cae-82b1-4da4-8785-29f9ddb1b05bn%40googlegroups.com.
Clea Smith
May 23, 2022, 7:36:58 AM5/23/22
to SpiNNaker Users Group
Hi Mirko,
I have done this replication on my local SpiNNaker board before, I would suggest you build your own neuron model if you could. Cus the threshold not only increases with each spike but also decays over long term.
It is fairly straightforward and easy to do.
I may not be able to disclose the detail of my code due to my NDA signed, But I do have pieces of my code on GitHub if it helps
Shouyu
mirkosan
May 24, 2022, 3:44:56 AM5/24/22
to SpiNNaker Users Group
Thank you for your reply.
I'm working with this new neuron model and I will check if it works for me.
Thanks Shouyu for your suggestion. I can try to create a neuron model but can you explain me (without violating your NDA) how to create it?
if you have free time to help me, I can send you my contacts.
Mirko
Andrew Gait
May 24, 2022, 3:56:59 AM5/24/22
to mirkosan, SpiNNaker Users Group
Hi Mirko,
You can read up on how to add new neuron models here:
http://spinnakermanchester.github.io/spynnaker/6.0.0/NewNeuronModels6.0-LabManual.pdf
I think it ought to be as straightforward as taking the C_model/Adap_threshold_type.h file from Shouyu's github link and adding it to the build structure for your new model. But if you have any questions, then please ask them.
Andy
----------------------------------------------------
Dr. Andrew Gait (he/him),
Dr. Andrew Gait (he/him),
Research Software Engineer,
APT Group,
Department of Computer Science,
The University of Manchester,
Oxford Road, Manchester M13 9PL
email: andre...@manchester.ac.uk
web: http://personalpages.manchester.ac.uk/staff/Andrew.Gait/
Department of Computer Science,
The University of Manchester,
Oxford Road, Manchester M13 9PL
email: andre...@manchester.ac.uk
web: http://personalpages.manchester.ac.uk/staff/Andrew.Gait/
I do not expect a reply to this email outside of normal working hours
From: spinnak...@googlegroups.com <spinnak...@googlegroups.com> on behalf of mirkosan <mirk...@hotmail.it>
Sent: 24 May 2022 08:44
Sent: 24 May 2022 08:44
Subject: Re: [SpiNNaker Mailing List] Adaptive treshold
To view this discussion on the web, visit
https://groups.google.com/d/msgid/spinnakerusers/ea4978c8-9b30-4e8e-b98e-8f315ea3d5e9n%40googlegroups.com.
mirkosan
May 24, 2022, 9:55:28 AM5/24/22
to SpiNNaker Users Group
Hi Andy,
I'm trying to follow the guide you sent me, it is not so easy for me but maybe I need more time to understand.
I have also some questions about the classification phase.
I don't know how to set labels after training to perform the classification.
I have tried also to find a solution to set the "learning rate" equal to zero for the classification phase, to make the STDP weights fixed, but I didn't find nothing in the documentation.
Maybe Shouyu could help me.
Thank you for your help.
Mirko
mirkosan
May 30, 2022, 4:52:41 AM5/30/22
to SpiNNaker Users Group
Hi all,
I tried to follow the guide.
I downloaded the zip file and I modified "my_threshold_type.h" file in \sPyNNaker8NewModelTemplate-6.0.0\c_models\src\my_models\threshold_types using the Adap_threshold_type.h file from Shouyu.
Then i run
python setup.py develop in cmd.
Are there other steps I have to do?
How can I add this new model on my collab to run it on SpiNNaker through Jupyter Notebook?
Thanks.
Mirko
Andrew Gait
May 30, 2022, 6:37:57 AM5/30/22
to mirkosan, SpiNNaker Users Group
Hi Mirko,
Are you running just using the latest release version (i.e. have you just done pip install sPyNNaker), or using the sPyNNaker kernel on Jupyter? Or are you using a recent version of master (via git) or the sPyNNakerGit kernel on Jupyter?
If you've just done pip install sPyNNaker, then you need to follow the instructions at
then replace the relevant file as you have done, and make sure all code is (re)built once you have replaced the relevant file.
If you're using a recent version of master then make sure you have sPyNNaker8NewModelTemplate cloned, replace the relevant .h file as you have done and then rebuild the C code using "make" in the c_models directory of sPyNNaker8NewModelTemplate.
And to see how to use the new model in action, look at the example in sPyNNaker8NewModelTemplate/examples/my_example.py
Any further questions, please ask.
Andy
----------------------------------------------------
Dr. Andrew Gait (he/him),
Dr. Andrew Gait (he/him),
Research Software Engineer,
APT Group,
Department of Computer Science,
The University of Manchester,
Oxford Road, Manchester M13 9PL
email: andre...@manchester.ac.uk
web: http://personalpages.manchester.ac.uk/staff/Andrew.Gait/
Department of Computer Science,
The University of Manchester,
Oxford Road, Manchester M13 9PL
email: andre...@manchester.ac.uk
web: http://personalpages.manchester.ac.uk/staff/Andrew.Gait/
I do not expect a reply to this email outside of normal working hours
From: spinnak...@googlegroups.com <spinnak...@googlegroups.com> on behalf of mirkosan <mirk...@hotmail.it>
Sent: 30 May 2022 09:52
To view this discussion on the web, visit
https://groups.google.com/d/msgid/spinnakerusers/517c5e4f-1dd5-4d75-9b03-54cb6f645d22n%40googlegroups.com.
mirkosan
May 30, 2022, 6:51:43 AM5/30/22
to SpiNNaker Users Group
Hi Andy,
I'm working on Ebrain Collaboratory -
SpiNNaker interactive on Jupyter. I'm using the SpyNNaker kernel.
I have a folder on Jupyter: sPyNNakerGit/sPyNNaker8NewModelTemplate. I have to modify the file in this folder?
Thanks
Mirko
Andrew Gait
May 30, 2022, 9:40:47 AM5/30/22
to mirkosan, SpiNNaker Users Group
Hi Mirko,
That's one way you can do it, yes, but then you will have to use the sPyNNakerGit kernel. Once you have edited the file you will also need to rebuild the C code - the easiest way to do this is to go into the sPyNNakerGit directory in a terminal window from
Jupyter and from there run the commands "source bin/activate" and then "bash support/automatic_make.sh". After a few minutes running the second of these commands this should compile everything you need for use in a notebook on Jupyter (again, don't forget
to use the sPyNNakerGit kernel).
Andy
----------------------------------------------------
Dr. Andrew Gait (he/him),
Dr. Andrew Gait (he/him),
Research Software Engineer,
APT Group,
Department of Computer Science,
The University of Manchester,
Oxford Road, Manchester M13 9PL
email: andre...@manchester.ac.uk
web: http://personalpages.manchester.ac.uk/staff/Andrew.Gait/
Department of Computer Science,
The University of Manchester,
Oxford Road, Manchester M13 9PL
email: andre...@manchester.ac.uk
web: http://personalpages.manchester.ac.uk/staff/Andrew.Gait/
I do not expect a reply to this email outside of normal working hours
From: spinnak...@googlegroups.com <spinnak...@googlegroups.com> on behalf of mirkosan <mirk...@hotmail.it>
Sent: 30 May 2022 11:51
To view this discussion on the web, visit
https://groups.google.com/d/msgid/spinnakerusers/69973d15-f8e6-4e15-b981-1f2bdcb6a19en%40googlegroups.com.
mirkosan
May 31, 2022, 4:21:56 AM5/31/22
to SpiNNaker Users Group
Hi Andy,
Thank you for your help. I followed all the steps and now, using a sPyNNakerGit kernel, I am able to import the new neuron model.
I still have some problems. If I try to set some parameters (like "threshold_value"), I got this error: "__init__() got an unexpected keyword argument 'threshold_value'.
The steps I have done are this:
I modified the file "my_threshold_type.h" stored in sPyNNakerGit/sPyNNaker8NewModelTemplate/c_models/src/my_models/threshold_types/ using the file Adap_threshold_type.h from Shouyu's GitHub.
Then, in a terminal, I run the commands "source bin/activate" and then "bash support/automatic_make.sh" as you suggested.
Then using "from python_models8.neuron.builds.my_model_curr_exp import MyModelCurrExp", I imported the new model on a SpyNNakerGit kernel.
Probably I miss some steps, maybe I have to modify the file "my_model_curr_exp.py".
I'm so sorry but I have no experiences on it and it's my first time with SpiNNaker and Spiking Neural Networks.
Thank you again for your help.
Mirko
Andrew Gait
May 31, 2022, 4:45:23 AM5/31/22
to mirkosan, SpiNNaker Users Group
Hi Mirko,
Ah - sorry, I probably should have explained this part too. You need to use a model that incorporates the "my_threshold_type.h" that you've just changed. But the only example that has that in sPyNNaker8NewModelTemplate also uses the neuron model "my_neuron_model_impl.h",
which I suspect you don't want to use and you want to use the standard LIF model instead? This involves making a new directory in the "makefiles" directory and a new equivalent build in the python code.
For the makefile: copy the directory my_model_curr_exp_my_threshold to if_curr_exp_my_threshold (or another name if you prefer), and in the new directory edit the Makefile, changing the two lines:
NEURON_MODEL = $(EXTRA_SRC_DIR)/my_models/models/my_neuron_model_impl.c
NEURON_MODEL_H = $(EXTRA_SRC_DIR)/my_models/models/my_neuron_model_impl.h
NEURON_MODEL_H = $(EXTRA_SRC_DIR)/my_models/models/my_neuron_model_impl.h
to
NEURON_MODEL = $(NEURON_DIR)/neuron/models/neuron_model_lif_impl.c
NEURON_MODEL_H = $(NEURON_DIR)/neuron/models/neuron_model_lif_impl.h
NEURON_MODEL_H = $(NEURON_DIR)/neuron/models/neuron_model_lif_impl.h
Then in python_models8/neuron/builds, copy my_model_curr_exp_my_threshold.py to if_curr_exp_my_threshold.py, and in this file you need to make sure that you're now using "NeuronModelLeakyIntegrateAndFire" rather than "MyNeuronModel", and that you're passing
the correct parameters into it (have a look at e.g. my_if_curr_exp_sEMD.py) for what these parameters are. Remember too to change the class name and the model name, probably to "IFCurrExpMyThreshold", and the binary name to match the name of the makefile
directory you set earlier (i.e. "if_curr_exp_my_threshold.aplx").
Then to use your new build in a script you would import it:
from python_models8.neuron.builds.if_curr_exp_my_threshold import (
IFCurrExpMyThreshold)
IFCurrExpMyThreshold)
This should then allow you to do everything you'd normally do in a script with regard to setting the relevant parameters.
I think that's everything you'd need to change. If you still have problems then try following examples 1 and 2 at the end of http://spinnakermanchester.github.io/spynnaker/6.0.0/NewNeuronModels6.0-LabManual.pdf
.
Do ask if you have any further problems that aren't explained by the lab manual or this email.
Andy
----------------------------------------------------
Dr. Andrew Gait (he/him),
Dr. Andrew Gait (he/him),
Research Software Engineer,
APT Group,
Department of Computer Science,
The University of Manchester,
Oxford Road, Manchester M13 9PL
email: andre...@manchester.ac.uk
web: http://personalpages.manchester.ac.uk/staff/Andrew.Gait/
Department of Computer Science,
The University of Manchester,
Oxford Road, Manchester M13 9PL
email: andre...@manchester.ac.uk
web: http://personalpages.manchester.ac.uk/staff/Andrew.Gait/
I do not expect a reply to this email outside of normal working hours
From: spinnak...@googlegroups.com <spinnak...@googlegroups.com> on behalf of mirkosan <mirk...@hotmail.it>
Sent: 31 May 2022 09:21
To view this discussion on the web, visit
https://groups.google.com/d/msgid/spinnakerusers/12435e47-e20d-44f4-b677-c9d46b8da7a6n%40googlegroups.com.
mirkosan
May 31, 2022, 9:58:20 AM5/31/22
to SpiNNaker Users Group
Hi Andy,
I followed all the steps but i got again an error:
ImportError: cannot import name 'NeuronModelLeakyIntegrateAndFire' from 'python_models8.neuron.neuron_models.my_neuron_model' (/home/bbpnrsoa/sPyNNakerGit/sPyNNaker8NewModelTemplate/python_models8/neuron/neuron_models/my_neuron_model.py)
In the attached file you can find a screenshot of the error and the if_curr_exp_my_threshold.py I created.
Thanks.
Mirko
Andrew Gait
May 31, 2022, 10:59:00 AM5/31/22
to mirkosan, SpiNNaker Users Group
Hi Mirko,
That should be
from spynnaker.pyNN.models.neuron.neuron_models import NeuronModelLeakyIntegrateAndFire
instead of what you have on line 7.
Andy
----------------------------------------------------
Dr. Andrew Gait (he/him),
Dr. Andrew Gait (he/him),
Research Software Engineer,
APT Group,
Department of Computer Science,
The University of Manchester,
Oxford Road, Manchester M13 9PL
email: andre...@manchester.ac.uk
web: http://personalpages.manchester.ac.uk/staff/Andrew.Gait/
Department of Computer Science,
The University of Manchester,
Oxford Road, Manchester M13 9PL
email: andre...@manchester.ac.uk
web: http://personalpages.manchester.ac.uk/staff/Andrew.Gait/
I do not expect a reply to this email outside of normal working hours
From: spinnak...@googlegroups.com <spinnak...@googlegroups.com> on behalf of mirkosan <mirk...@hotmail.it>
Sent: 31 May 2022 14:58
To view this discussion on the web, visit
https://groups.google.com/d/msgid/spinnakerusers/f942a34e-eaf5-4f34-bed2-ede36e6c4cc4n%40googlegroups.com.
mirkosan
Jun 1, 2022, 4:34:40 AM6/1/22
to SpiNNaker Users Group
Hi Andy,
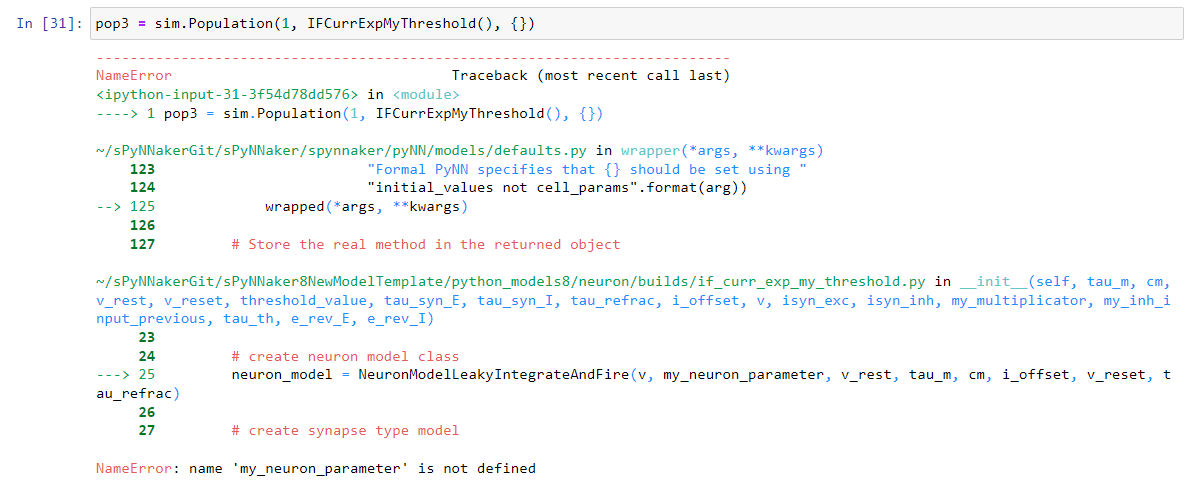
I understand that this conversation is getting too long and I hope you don't get too much trouble answering every question I have.
I modified the line 7 and now I have no errors importing the new model but when I create the population I got the error in the screenshot.
It is strange because in "if_curr_exp_my_threshold.py" I defined the parameter "my_neuron_parameter".
In the attached file you can find the file "if_curr_exp_my_treshold.py"
Thanks
Mirko
Andrew Gait
Jun 1, 2022, 5:27:11 AM6/1/22
to mirkosan, SpiNNaker Users Group
Hi Mirko,
The Manchester mail server doesn't like attached .py files so we can't see your attachment - can you zip it and send it in zip form?
Thanks,
Andy
----------------------------------------------------
Dr. Andrew Gait (he/him),
Dr. Andrew Gait (he/him),
Research Software Engineer,
APT Group,
Department of Computer Science,
The University of Manchester,
Oxford Road, Manchester M13 9PL
email: andre...@manchester.ac.uk
web: http://personalpages.manchester.ac.uk/staff/Andrew.Gait/
Department of Computer Science,
The University of Manchester,
Oxford Road, Manchester M13 9PL
email: andre...@manchester.ac.uk
web: http://personalpages.manchester.ac.uk/staff/Andrew.Gait/
I do not expect a reply to this email outside of normal working hours
From: spinnak...@googlegroups.com <spinnak...@googlegroups.com> on behalf of mirkosan <mirk...@hotmail.it>
Sent: 01 June 2022 09:34
To view this discussion on the web, visit
https://groups.google.com/d/msgid/spinnakerusers/92b095d3-c389-4bb2-bb61-56134eae4c52n%40googlegroups.com.
Andrew Gait
Jun 1, 2022, 10:05:03 AM6/1/22
to mirkosan, SpiNNaker Users Group
Thanks Mirko,
The file isn't quite right - the arguments aren't correct for some of the model components. I think you need something like this instead:
def __init__(
self,
tau_m=20.0, cm=1.0, v_rest=-65.0, v_reset=-65.0,
threshold_value=-52.0, tau_syn_E=5.0, tau_syn_I=5.0, tau_refrac=0.1,
i_offset=0.0, v=-65.0, isyn_exc=0.0, isyn_inh=0.0,
e_rev_E=0, e_rev_I= -100, tau_th= (10 ** 5)*5, theta=0.5):
# create neuron model class
neuron_model = NeuronModelLeakyIntegrateAndFire(v, v_rest, tau_m, cm, i_offset, v_reset, tau_refrac)
# create synapse type model
synapse_type = SynapseTypeExponential(
tau_syn_E, tau_syn_I, isyn_exc, isyn_inh)
# create input type model
input_type = InputTypeCurrent()
# create threshold type model
threshold_type = MyThresholdType(
threshold_value, tau_th, theta)
You also need to make sure you've edited python_models8/neuron/threshold_types/my_threshold_type.py correctly in order to add the tau_th and theta alongside the threshold_value parameter (and I think remove the my_threshold_parameter). Make sure that the values
you are setting on the python side for this match the values in the struct in the C code you have added (in this case threshold_value, tau_th and theta). I'm not sure what the default values for these parameters are - you may need to check that with Clea
or have a look at her github python code to work it out.
Don't worry if you feel like you're asking a lot of questions - we want to get this working for you.
Andy
----------------------------------------------------
Dr. Andrew Gait (he/him),
Dr. Andrew Gait (he/him),
Research Software Engineer,
APT Group,
Department of Computer Science,
The University of Manchester,
Oxford Road, Manchester M13 9PL
email: andre...@manchester.ac.uk
web: http://personalpages.manchester.ac.uk/staff/Andrew.Gait/
Department of Computer Science,
The University of Manchester,
Oxford Road, Manchester M13 9PL
email: andre...@manchester.ac.uk
web: http://personalpages.manchester.ac.uk/staff/Andrew.Gait/
I do not expect a reply to this email outside of normal working hours
From: spinnak...@googlegroups.com <spinnak...@googlegroups.com> on behalf of mirkosan <mirk...@hotmail.it>
Sent: 01 June 2022 11:57
To view this discussion on the web, visit
https://groups.google.com/d/msgid/spinnakerusers/0cf46d73-5187-4726-98bb-f673e056bd21n%40googlegroups.com.
Message has been deleted
Message has been deleted
Message has been deleted
Message has been deleted
Andrew Gait
Jun 9, 2022, 4:35:05 AM6/9/22
to mirkosan, SpiNNaker Users Group
Hi Mirko,
Ah - sorry, I think that's partly my fault. In the suggestion I sent you, you shouldn't have the parameters "e_rev_E" or "e_rev_I". If you delete these from the list inside the init(..) then this should now work.
(As a note for the future: all the parameters in the init(..) of a new model should then be used in the components in that function; if they are not then that can lead to problems like the ones you are seeing).
Andy
----------------------------------------------------
Dr. Andrew Gait (he/him),
Dr. Andrew Gait (he/him),
Research Software Engineer,
APT Group,
Department of Computer Science,
The University of Manchester,
Oxford Road, Manchester M13 9PL
email: andre...@manchester.ac.uk
web: http://personalpages.manchester.ac.uk/staff/Andrew.Gait/
Department of Computer Science,
The University of Manchester,
Oxford Road, Manchester M13 9PL
email: andre...@manchester.ac.uk
web: http://personalpages.manchester.ac.uk/staff/Andrew.Gait/
I do not expect a reply to this email outside of normal working hours
From: spinnak...@googlegroups.com <spinnak...@googlegroups.com> on behalf of mirkosan <mirk...@hotmail.it>
Sent: 07 June 2022 14:58
To: SpiNNaker Users Group <spinnak...@googlegroups.com>
Subject: Re: [SpiNNaker Mailing List] Adaptive treshold
Hi Andy,
I followed your suggestions and now I can set all the parameters but when I try to retrieve the spikes through "neo = pop.get_data(variables=["spikes", "v"])" I got this error:
InvalidParameterType: Population IFCurrExpMyThreshold does not have parameter e_rev_E
Maybe I have to modify also the file "population.py"?
Thanks
Mirko
From: spinnakerusers@googlegroups.com <spinnakerusers@googlegroups.com> on behalf of mirkosan <mirk...@hotmail.it>
Sent: 01 June 2022 11:57
To unsubscribe from this group and stop receiving emails from it, send an email to spinnakerusers+unsubscribe@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/spinnakerusers/0cf46d73-5187-4726-98bb-f673e056bd21n%40googlegroups.com.
--
You received this message because you are subscribed to the Google Groups "SpiNNaker Users Group" group.
To unsubscribe from this group and stop receiving emails from it, send an email to spinnakeruser...@googlegroups.com.
You received this message because you are subscribed to the Google Groups "SpiNNaker Users Group" group.
To unsubscribe from this group and stop receiving emails from it, send an email to spinnakeruser...@googlegroups.com.
To view this discussion on the web, visit
https://groups.google.com/d/msgid/spinnakerusers/a9a22db5-abf0-4cc5-9a12-14e9d1b64f0fn%40googlegroups.com.
Message has been deleted
Andrew Gait
Jun 9, 2022, 8:36:04 AM6/9/22
to mirkosan, SpiNNaker Users Group, Clea Smith
Hi Mirko,
As it says in the error message, that suggests that you are trying to set a particular value (looking at your variable list, it's tau_th) to 500000.0. The 32-bit fixed point numbers we have to use on SpiNNaker (as we don't have hardware floating point) do
not allow you to use a value this large.
I suspect what has happened here is that in the other user's example, the actual value sent to the machine is not the tau_th value itself, but an exponential calculated from that value. As I can't see their actual python code for doing this, I don't know exactly
what they used, but I suspect that it was something along the lines of exp_th = exp(-(1/tau_th)), or perhaps it's simply just exp_th = 1/tau_th, I'm not sure. To replicate this you'll need to edit your neuron/threshold_types/my_threshold_type.py, and change
anything that says parameters[TAU_TH] in the get_values(...) function to whatever you want to do - e.g. to
parameters[TAU_TH].apply_operation(lambda x: 1.0 / x)
if you want to do 1/tau_th, or
parameters[TAU_TH].apply_operation(lambda x: numpy.exp(-1.0/x))
if you want to do exp(-1/tau_th). Note that in this case you'll also have to add an "import numpy" as a line at the top of my_threshold_type.py.
If you're still having problems then send over your my_threshold_type.py file (zipped) and we can go from there.
Perhaps Clea could also help us out here by providing their code for what they did... ?
Andy
----------------------------------------------------
Dr. Andrew Gait (he/him),
Dr. Andrew Gait (he/him),
Research Software Engineer,
APT Group,
Department of Computer Science,
The University of Manchester,
Oxford Road, Manchester M13 9PL
email: andre...@manchester.ac.uk
web: http://personalpages.manchester.ac.uk/staff/Andrew.Gait/
Department of Computer Science,
The University of Manchester,
Oxford Road, Manchester M13 9PL
email: andre...@manchester.ac.uk
web: http://personalpages.manchester.ac.uk/staff/Andrew.Gait/
I do not expect a reply to this email outside of normal working hours
From: spinnak...@googlegroups.com <spinnak...@googlegroups.com> on behalf of mirkosan <mirk...@hotmail.it>
Sent: 09 June 2022 09:53
To: SpiNNaker Users Group <spinnak...@googlegroups.com>
Subject: Re: [SpiNNaker Mailing List] Adaptive treshold
Hi Andy,
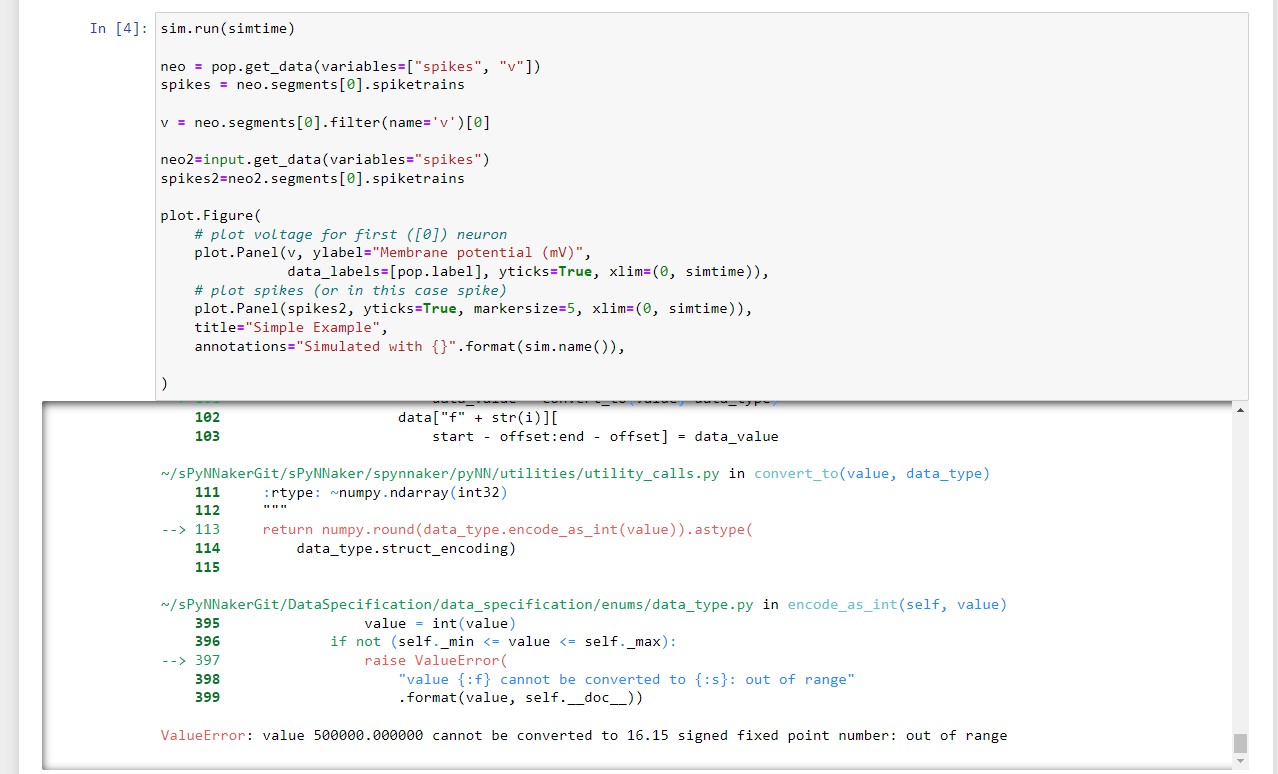
I modified it and I got another error message when I tried to run the simulation
Thanks
Mirko
Il giorno giovedì 9 giugno 2022 alle 10:35:05 UTC+2 Andrew Gait ha scritto:
Hi Mirko,
From: spinnak...@googlegroups.com <spinnak...@googlegroups.com> on behalf of mirkosan <mirk...@hotmail.it>
Sent: 01 June 2022 11:57
To: SpiNNaker Users Group <spinnak...@googlegroups.com>
Subject: Re: [SpiNNaker Mailing List] Adaptive treshold
Hi Andy,
To view this discussion on the web, visit https://groups.google.com/d/msgid/spinnakerusers/0cf46d73-5187-4726-98bb-f673e056bd21n%40googlegroups.com.
--
You received this message because you are subscribed to the Google Groups "SpiNNaker Users Group" group.
To unsubscribe from this group and stop receiving emails from it, send an email to spinnakeruser...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/spinnakerusers/a9a22db5-abf0-4cc5-9a12-14e9d1b64f0fn%40googlegroups.com.
--
You received this message because you are subscribed to the Google Groups "SpiNNaker Users Group" group.
To unsubscribe from this group and stop receiving emails from it, send an email to spinnakeruser...@googlegroups.com.
You received this message because you are subscribed to the Google Groups "SpiNNaker Users Group" group.
To unsubscribe from this group and stop receiving emails from it, send an email to spinnakeruser...@googlegroups.com.
To view this discussion on the web, visit
https://groups.google.com/d/msgid/spinnakerusers/fc43304c-0f75-4b24-8eaa-b589134b4df8n%40googlegroups.com.
Reply all
Reply to author
Forward
Message has been deleted
0 new messages
