command "C:/TPP/bin/PeptideProphetParser" failed
46 views
Skip to first unread message
Asif Ahmed
Dec 2, 2020, 1:27:44 PM12/2/20
to spctools-discuss
Hi,
![]()
I did a comet search (Comet version "2020.01 rev. 0") in Linux HPC, update the paths of .mzML files, but when I tried to combine and run peptide-prophet (on TPP v5.2.0), I am getting this error. I tried to combine all the files, without running peptide-prophet and it works!
Can you please advice me- what to do?
ASIF
Jimmy Eng
Dec 2, 2020, 1:50:02 PM12/2/20
to spctools...@googlegroups.com
Stick with Comet 2019.01.5. In version 2020.01.0, I deprecated the "deltacnstar" score in Comet and it looks like PeptideProphet can't run w/o it. Alternately, run this Perl command on your pep.xml files before running PeptideProphet if you want a workaround with the latest Comet:
perl -i -pe 's/<search_score name="deltacn"/<search_score name="deltacnstar" value="0.0"\/>\n <search_score name="deltacn"/g;' yourfile.pep.xml
--
You received this message because you are subscribed to the Google Groups "spctools-discuss" group.
To unsubscribe from this group and stop receiving emails from it, send an email to spctools-discu...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/spctools-discuss/2537e476-9ae1-45c4-974d-c087a337c667n%40googlegroups.com.
David Shteynberg
Dec 2, 2020, 1:58:52 PM12/2/20
to spctools-discuss
I have updated PeptideProphetParser so that it CAN run with the new version of Comet. You will have to get it from the TPP SVN repository.
To view this discussion on the web visit https://groups.google.com/d/msgid/spctools-discuss/CAJqD6EN9s1Dbiu1eSkcq64A4%3DG_T_%3DRoqeyqgR98iGJcR9u04g%40mail.gmail.com.
Asif Ahmed
Dec 2, 2020, 2:23:45 PM12/2/20
to spctools...@googlegroups.com
Thanks Jimmy, i will try your suggestion. It took a while to generate the pep.xml files, So I would prefer your perl script.
David, is it possible to share the link? i was going to source forge website, but could not find it.
You received this message because you are subscribed to a topic in the Google Groups "spctools-discuss" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/spctools-discuss/TXNSCJwoGuU/unsubscribe.
To unsubscribe from this group and all its topics, send an email to spctools-discu...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/spctools-discuss/CAGJJY%3D8XDEEXaGUGV8cWaeeHwNNB3SeAeGCaR6mygF-FB_mR-w%40mail.gmail.com.
Khandaker Asif Ahmed
PhD Student| Applied BioSciences, Macquarie University, NSW, Australia
Postgraduate Research Student| CSIRO Land and Water Flagship, Black Mountain, ACT, Australia
Address: Room No: S1.03, Building No: 101, CSIRO Clunies Ross Street, Black Mountain, ACT 2601, Australia
Mobile: (+61)0434018803, Skype ID: kh.asifratul
David Shteynberg
Dec 2, 2020, 2:30:49 PM12/2/20
to spctools-discuss
Are you building the TPP from source? All the code is here: https://sourceforge.net/p/sashimi/code/HEAD/tree/trunk/trans_proteomic_pipeline/ but if building from SVN source is too much headache we should have an official release coming up in the next few weeks give or take. Meanwhile, Jimmy's solution will also have the same result.
To view this discussion on the web visit https://groups.google.com/d/msgid/spctools-discuss/CABV_oz_%2BJin1n0WVdCrb%2BiqfPUj3Op5%3DiDH4SVgPq4KHabB%3D4Q%40mail.gmail.com.
Asif Ahmed
Dec 2, 2020, 2:41:50 PM12/2/20
to spctools...@googlegroups.com
I am using the windows version,
any suggestion for that?
To view this discussion on the web visit https://groups.google.com/d/msgid/spctools-discuss/CAGJJY%3D9Wz-z4BA1ujQrHik801_o%2BUibbdYJasFpp5OCxoU7Uyw%40mail.gmail.com.
David Shteynberg
Dec 2, 2020, 2:43:54 PM12/2/20
to spctools-discuss
Use Jimmy's solution until the next release.
To view this discussion on the web visit https://groups.google.com/d/msgid/spctools-discuss/CABV_oz-5Cd%3D0KhT-4dStif%2B15gczTH-pxt_FQ1iSA-m-4s7nkQ%40mail.gmail.com.
Asif Ahmed
Dec 2, 2020, 3:07:38 PM12/2/20
to spctools...@googlegroups.com
The script works perfectly without any error! Thanks for your help.
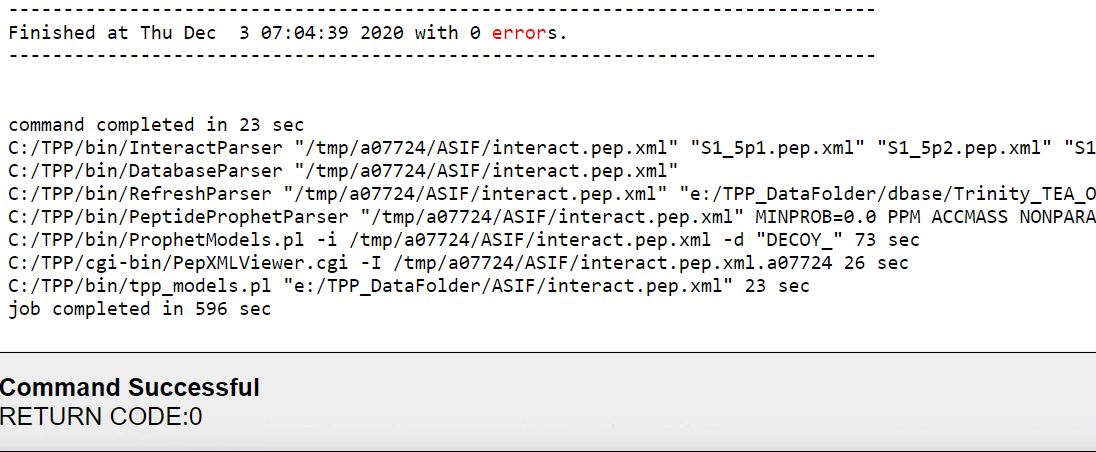

To view this discussion on the web visit https://groups.google.com/d/msgid/spctools-discuss/CAGJJY%3D8xJzicksxYT%3DjRQXbU7Hqq5u5dNxg5nmK_HspEJ%2BPTdg%40mail.gmail.com.
Reply all
Reply to author
Forward
0 new messages
