Soft sweep simulation too slow
Anushka
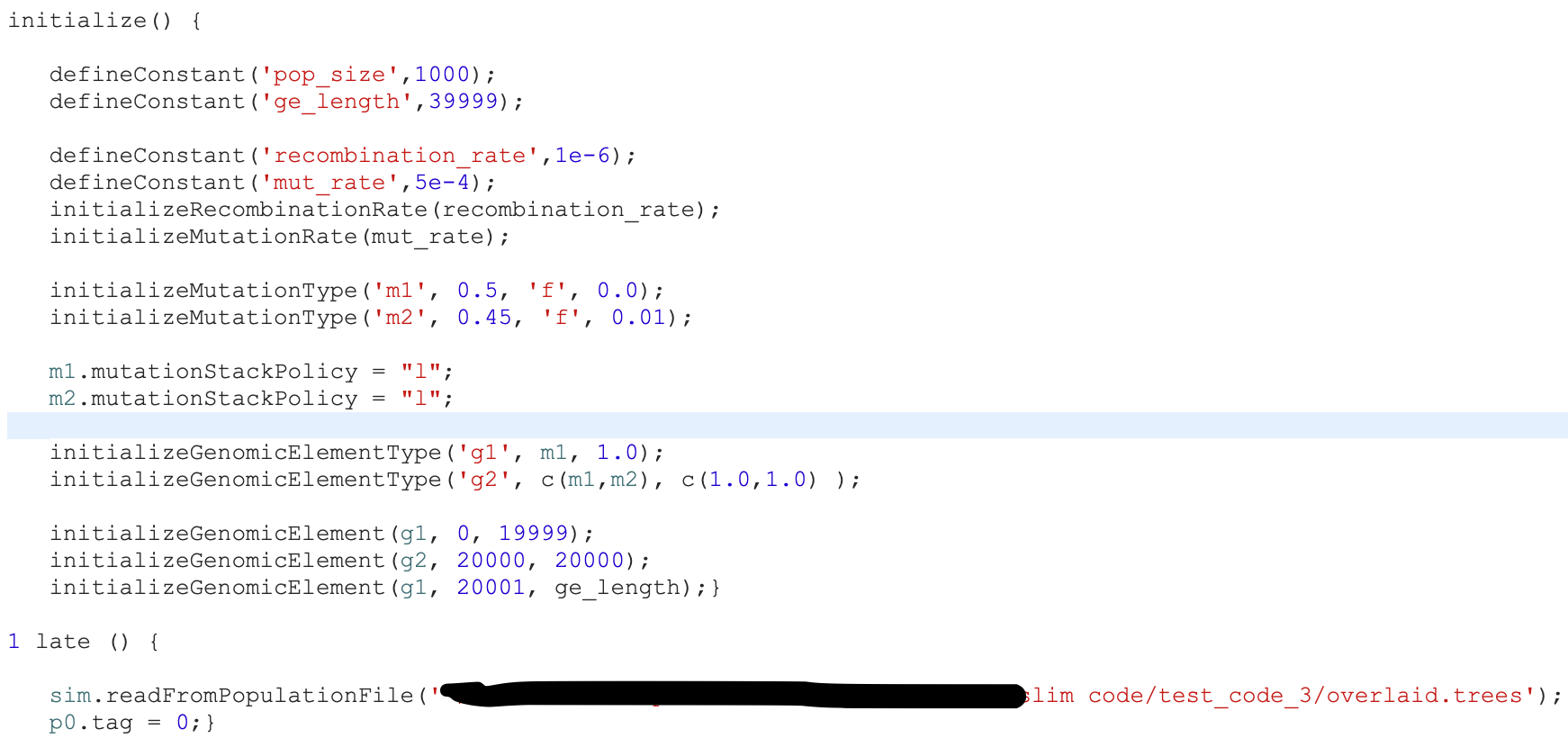
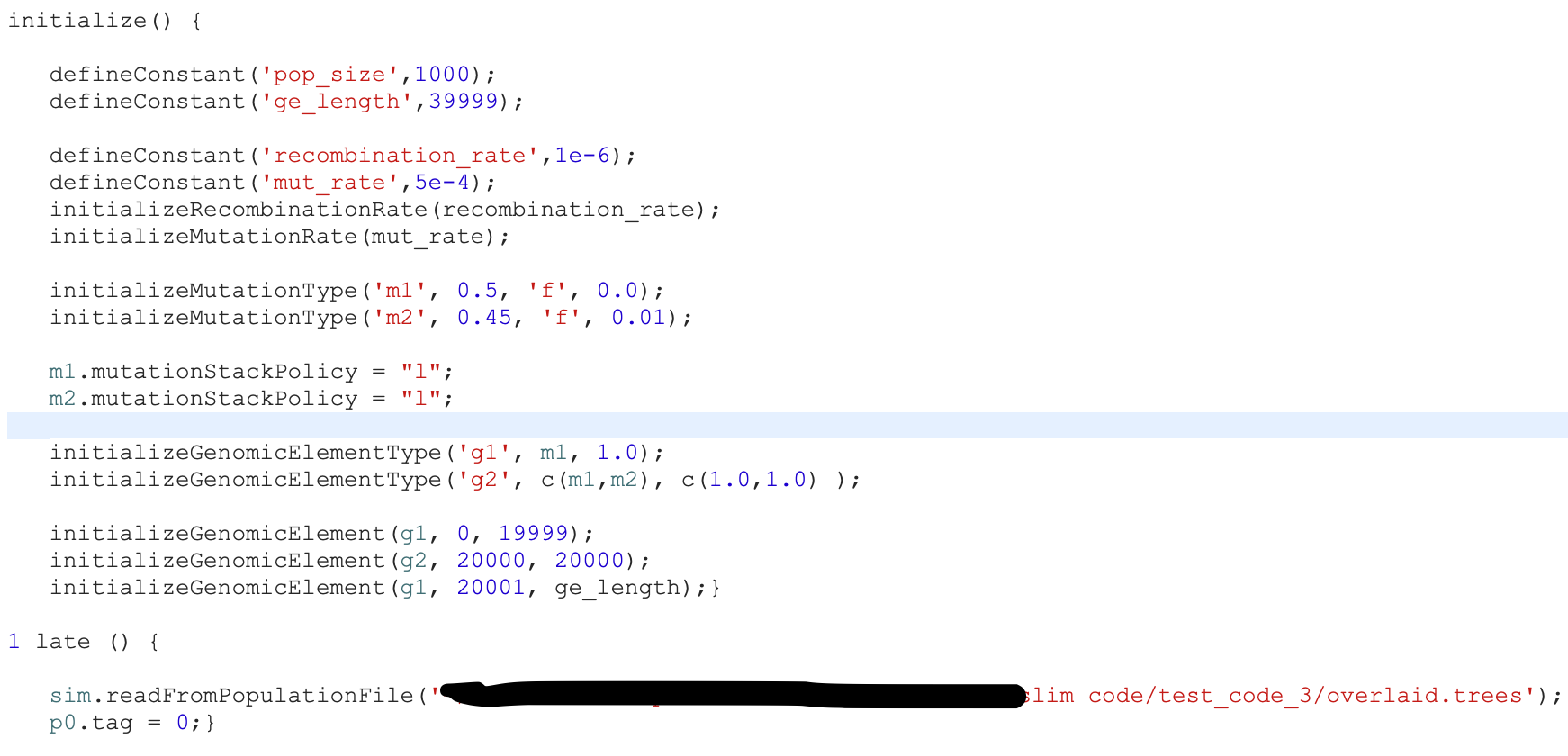
initialize() {
defineConstant('simID', getSeed());
defineConstant('pop_size',10000);
defineConstant('ge_length',99999);
defineConstant('sweep_dom',0.45);
defineConstant('recombination_rate',1e-3);
defineConstant('mut_rate',1/(4*pop_size));
initializeRecombinationRate(recombination_rate);
initializeMutationRate(mut_rate);
// Create two neutral mutations. m2 will be the sweep mutation
initializeMutationType('m1', 0.5, 'f', 0.0);
initializeMutationType('m2', sweep_dom, 'f', 0.0);
m2.convertToSubstitution = F; // (is this needed?)
// Initialise genomic element with resistance mutation at position 50000
initializeGenomicElementType('g1', m1, 1.0 );
initializeGenomicElementType('g2', c(m1,m2), c(1.0,1.0) );
initializeGenomicElement(g1, 0, 49999);
initializeGenomicElement(g2, 50000, 50000);
initializeGenomicElement(g1, 50001, ge_length); }
1 {
sim.addSubpop('p0', pop_size);
p0.tag = 0;
start = clock(); }
2:99000 late() {
if (sim.generation % 500 == 0) {
catn(sim.generation);}
if (sim.generation % 3000 == 0){
catn("Elapsed: " + (clock() - start));}
}
100000 late() {
sim.outputFull(filePath = "slim_" + simID + ".txt");
sim.simulationFinished();
}
PART 2: Soft sweep
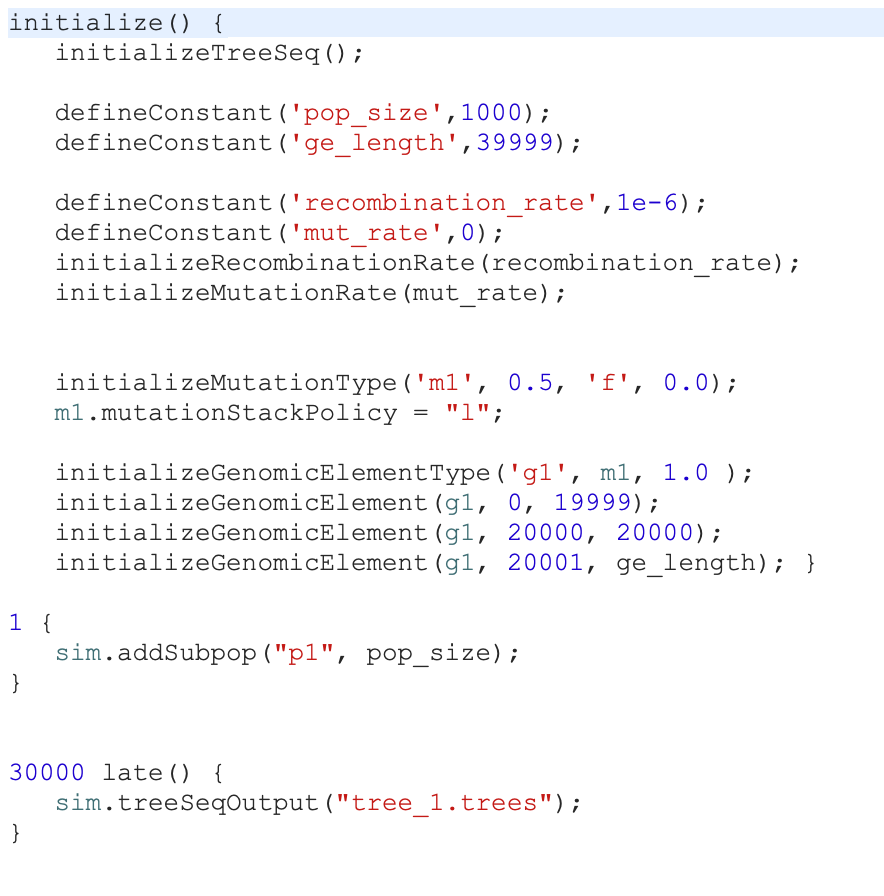
initialize() {
defineConstant('simID', getSeed());
defineConstant('pop_size',10000);
defineConstant('ge_length',99999);
defineConstant('sweep_dom',0.45);
defineConstant('recombination_rate',1e-3);
defineConstant('mut_rate',1/(4*pop_size));
initializeRecombinationRate(recombination_rate);
initializeMutationRate(mut_rate);
// Create two neutral mutations. m2 will be the sweep mutation
initializeMutationType('m1', 0.5, 'f', 0.0);
initializeMutationType('m2', sweep_dom, 'f', 0.0);
m2.convertToSubstitution = F; // (is this needed?)
// Initialise genomic element with resistance mutation at position 50000
initializeGenomicElementType('g1', m1, 1.0 );
initializeGenomicElementType('g2', c(m1,m2), c(1.0,1.0) );
initializeGenomicElement(g1, 0, 49999);
initializeGenomicElement(g2, 50000, 50000);
initializeGenomicElement(g1, 50001, ge_length); }
1 late () {
sim.readFromPopulationFile("~/Desktop/slim_" + '2353137633341' + ".txt");
p0.tag = 0;
}
2:5000 late() {
// Change selection coefficient of all m2 mutations to 0.01
sweep = sim.mutationsOfType(m2);
sweep.setSelectionCoeff(0.01);
if (p0.tag != sim.countOfMutationsOfType(m2)) {
if (any(sim.substitutions.mutationType == m2)) {
catn('HARD sweep ended in gen. ' + sim.generation);
sim.simulationFinished();}
else {
p0.tag = sim.countOfMutationsOfType(m2);
if (sim.generation % 100 == 0){
catn('Gen. ' + sim.generation + ': ' + p0.tag + ' lineage(s)');
catn('m2 frequency across all lineages: ' + sum(sim.mutationFrequencies(p0, sim.mutationsOfType(m2))));}
if ((p0.tag == 0) & (sim.generation > 3000)) {
catn('Sweep failed to establish');
sim.simulationFinished();}}}
if (all(p0.genomes.countOfMutationsOfType(m2) > 0)) {
catn('SOFT sweep ended in gen. ' + sim.generation);
catn('Frequencies: ');
print(sim.mutationFrequencies(p0, sim.mutationsOfType(m2))); // prints mutation frequency for each lineage
catn(p0.tag + 'lineages') ;
sample = p0.sampleIndividuals(300).genomes;
sample.outputVCF("soft_sweep_300_1.vcf");
sim.simulationFinished();}}
I would appreciate any guidance or advice.
Warm regards,
Anushka
Ben Haller
-B.
Benjamin C. Haller
Messer Lab
Cornell University
Ben Haller
-B.
Benjamin C. Haller
Messer Lab
Cornell University
Anushka
Thank you very much for your suggestions, I will try lowering the mutation and recombination rates first and see if that helps.
Anushka

Peter Ralph
Anushka
Anushka

Peter Ralph
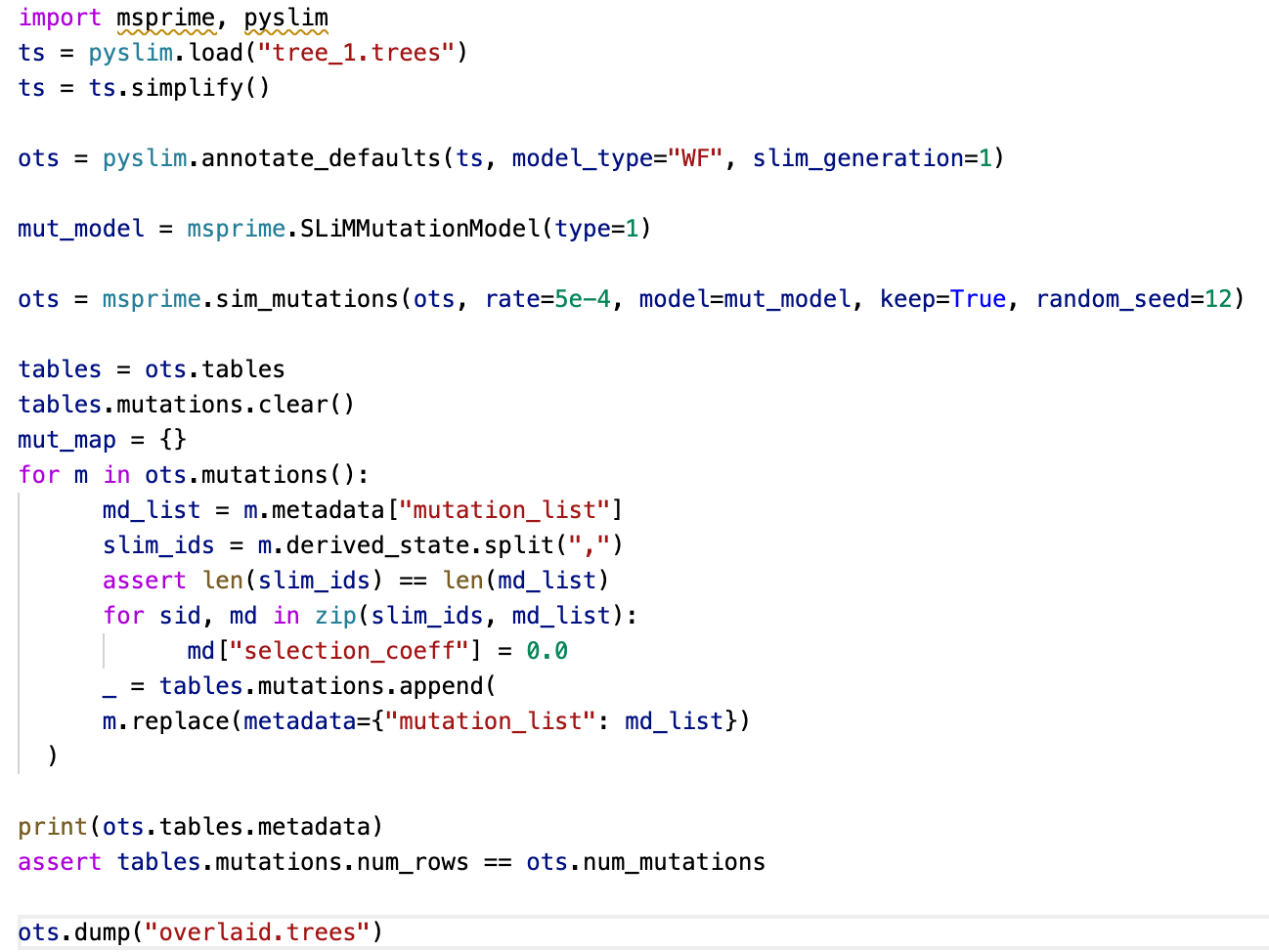
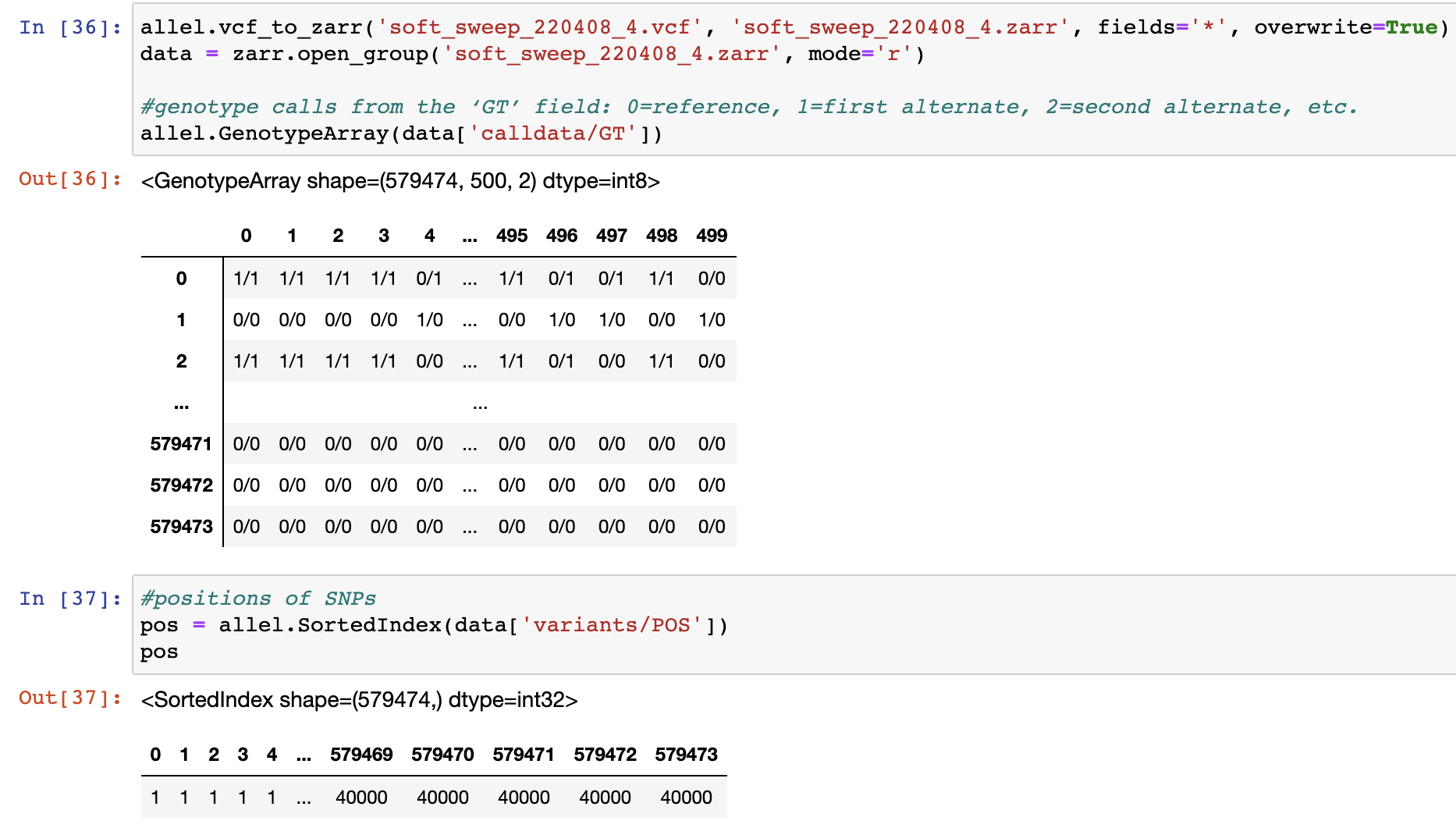
Thank you very much for your reply. I still don't understand why, given that I set the stacking policy to 'l', one individual (e.g. column 498 in the matrix below) can have two different alleles (alleles 0 and 2) at the same position (position 1). Could you please explain that?
Ben Haller
So, to resolve the issue you're having, you could (a) use a lower mutation rate to get fewer stacked mutations perhaps, (b) filter out sites with stacked mutations (but with your current mutation rate that might filter out almost all sites!), or (c) post-process the VCF data to determine the final mutation at each site, considering the most recent mutation in any stack to be the only mutation present. Perhaps (c) is your best option if you need the high mutation rate. It would be a custom script, in R or Python or C or whatever, that you would need to write, but perhaps it would not be too terribly difficult to get working. So then Python would still be overlaying the mutations with stacking, but you would essentially re-interpret the final data as if stacking were in effect. If necessary, you could also possibly do this in Python immediately after mutation overlay, fixing the actual tree sequence, but I have no idea how you'd do that or whether it would be difficult. Anyhow, if you do write such a script, I hope you will share it on slim-discuss or with a pull request to SLiM-Extras.
I guess that brings us to option (d): dive into the Python code for msprime/pyslim/tskit and add a new mutation stacking policy feature to it, parallel to how it works in SLiM. If you're up for that, perhaps Peter would have helpful advice regarding where to start. All of this stuff is open-source software, and is a community effort; you're welcome to join the community! :->
Cheers,
-B.
Benjamin C. Haller
Messer Lab
Cornell University
--
SLiM forward genetic simulation: http://messerlab.org/slim/
---
You received this message because you are subscribed to the Google Groups "slim-discuss" group.
To unsubscribe from this group and stop receiving emails from it, send an email to slim-discuss...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/slim-discuss/CAO0xWMD5aDT4o3y7e_8W8-HNAgJrunLWYpYVckLSn9HVyZE7sA%40mail.gmail.com.