select hits
13 views
Skip to first unread message
Michał T. Lorenc
Oct 21, 2021, 11:21:07 PM10/21/21
to sequenceserver
Hi.
We notice that it is possible to select only the best hit. Is there a way to select hit b?
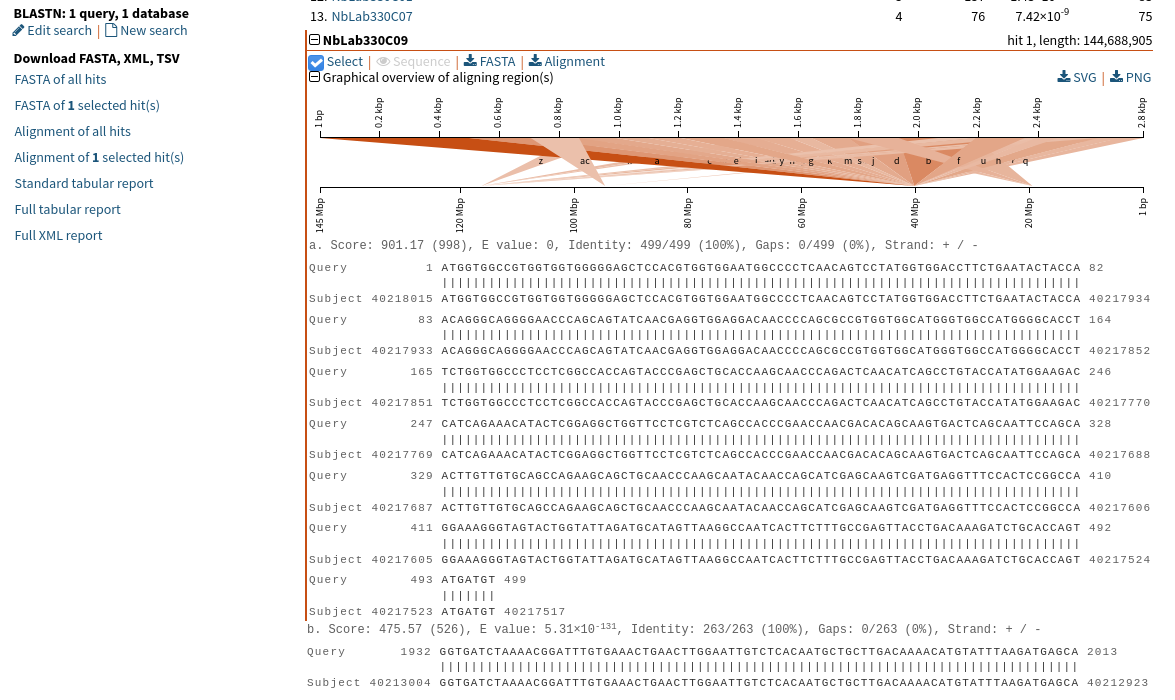
Thank you in advance,
Michal
Yannick Wurm
Oct 22, 2021, 12:26:00 PM10/22/21
to sequenceserver
Hi Michal,
thanks for the message. In the world of BLAST, "hit" means a sequence with one identifier in the database. In Sequenceserver, we number the hits. So that select button applies to that hit - i.e. NBlab330C09, and should download the entire sequence.
The letters refer to different HSPs ("high-scoring pairs" - ie. chunks that align between query and the hit sequence). So for your particular example, there are 29 or more such HSP. To differentiate HSPs from hits, in SequenceServer, we label them alphabetically a, b, c, and so-forth in the picture, but also in the pairwise alignments (your screenshot shows a. and the top of b.).
If you want to independently download all of HSPs, ie. the chunks of database sequence that aligns to your query sequences, you can click on the download Alignment button. That gives you a FASTA of all HSPs. The format is a bit funny, in that it includes the query sequence, "alignment strength", "hit sequence" (see my screenshot - where I added some whitespace to facilitate interpretation). I think you're looking for every third sequence in there.
Currently, those sequences are not identified using letters a, b, c, in the output. Instead, we show the from-to coordinates. I think we should have a, b, c listed and have opened an issue to that extent https://github.com/wurmlab/sequenceserver/issues/554
I hope this helps
Yannick
---
https://wurmlab.com - Evolutionary Genomics & Bioinformatics
Check https://sequenceserver.com/cloud - fast BLAST searches without the hassle of servers or upgrades
Reply all
Reply to author
Forward
0 new messages
