About model validation
598 views
Skip to first unread message
Bereket Workie Anteneh
Jul 12, 2021, 7:57:06 AM7/12/21
to scalismo
Dear Scalismo community,
I was built SSM of proximal femur with 24 training sets. I want to do model validation using PCA; (Generalization ability, specificity and compactness) of the model in Scalismo. Meanwhile, I was a little bit challenged to write the code for performing the validation.
So, would you have the code that simulate the validation via Generalization ability, specificity and compactness? Anybody who can share me the code is very appreciable.
With regards,
Marcel Luethi
Jul 13, 2021, 3:51:33 AM7/13/21
to Bereket Workie Anteneh, scalismo
Hi Bereket
The following code snippet computes the generalisation, specificity and the variance (which you need for computing compactness).
I hope this helps to get you started. You might also want to read the API doc of the individual methods to learn what the parameters mean.
Best regards,
Marcel
val reference : TriangleMesh[_3D] = ???
val trainingData : Seq[TriangleMesh[_3D]] = ???
val dataCollection : DataCollection[_3D, TriangleMesh, EuclideanVector[_3D]] = DataCollection.fromTriangleMesh3DSequence(reference, trainingData)
val model = PointDistributionModel.createUsingPCA(dataCollection)
val generalisation : Double = ModelMetrics.generalization(model, dataCollection).get
val specificity : Double = ModelMetrics.specificity(model, trainingData, 10)
// Variance captured in first 5 components
val variance : Double = breeze.linalg.sum(model.gp.variance(0 to 5))
--
You received this message because you are subscribed to the Google Groups "scalismo" group.
To unsubscribe from this group and stop receiving emails from it, send an email to scalismo+u...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/scalismo/d28b13b0-ab65-4c27-89f2-a461be5721bcn%40googlegroups.com.
Bereket Workie Anteneh
Jul 15, 2021, 8:25:40 AM7/15/21
to scalismo
Thank you Marcel for sharing what I want,
But still now I have a question specially in the code line 2 that is "val trainingData : Seq[TriangleMesh[_3D]] = ???"
The path of my training data was at "C:\\Users\\user\\Documents\\minimal-scalismo-seed-master\\data\\2Projecetd Meshes\\"
When I try to declare the path as shown below
val trainingData : Seq[TriangleMesh[_3D]] = MeshIO.readMesh(new File("C:\\Users\\user\\Documents\\minimal-scalismo-seed-master\\data\\2Projecetd Meshes\\")).get
It says an error, it says required: "Seq[TriangleMesh[_3D]] found: TriangleMesh [_3D]".
I tried a lot to fix it but it challenges me. So what would be the correct path declaration based on the training data directory that I mentioned above?
With regards,
Marcel Luethi
Jul 16, 2021, 10:39:18 PM7/16/21
to Bereket Workie Anteneh, scalismo
Hi Bereket
As the type signature says, you need a sequence of triangle meshes. To obtain such a sequence, you will need to read several meshes individually, typically within a for loop. The following code illustrates how this can be done. In the example, all files in a directory that end with .ply are read and returned as a sequence.
def readMeshesFromDirectory(directory : java.io.File) : Seq[TriangleMesh[_3D]] = {
val plyFiles = directory.listFiles().filter(file => file.getName.endsWith(".ply"))
for (plyFile <- plyFiles) yield {
MeshIO.readMesh(plyFile).get
}
}
readMeshesFromDirectory(new java.io.File("directoryContainingMeshes"))
Best regards,
Marcel
To view this discussion on the web visit https://groups.google.com/d/msgid/scalismo/d1bb0671-efc3-4814-8326-bc491e7a2bc0n%40googlegroups.com.
Bereket Workie Anteneh
Jul 18, 2021, 10:50:45 AM7/18/21
to Marcel Luethi, scalismo
Thank you Marcel, now it works well.
After I run the code you provide, the following numbers are seen in the result panel,
generalization 1.4810818231927857E-6
specificity 1.4436597052211486
variance 437794.1382516096
variance 437794.1382516096
What does these numbers standsfore/indicate?
With regards,
Marcel Luethi
Jul 19, 2021, 2:41:05 AM7/19/21
to Bereket Workie Anteneh, scalismo
Hi Bereket
The basic units in Scalismo is mm, which means you have a generaisation of 1.48e-6 mm (so almost 0) a specificity of 1.44 mm and a variance of 437794 mm^2
The metrics are described in depth in this paper:
Note that in the paper they describe compactness as the percentage of variance captured by the first n principal components. But once you know how to compute the variance this is easy to compute.
Best regards,
Marcel
Bereket Workie Anteneh
Jul 23, 2021, 8:30:55 AM7/23/21
to Marcel Luethi, scalismo
Thank you again Marcel,
From the code you provided " val specificity : Double = ModelMetrics.specificity(model, trainingData, 10)"
What does number 10 stand for? Is it changeable in each PC component/mode or is it taken as a constant?
Andreas Morel-Forster
Jul 27, 2021, 4:52:25 AM7/27/21
to scalismo
Hi Bereket
The number 10 indicates that the metric is calculated based on 10 samples. To get a more precise and reliable information, you might have to use there a higher number. See the documentation of the function attached below for more information.
/**
* Returns the specificity metric of the Statistical Mesh Model, that is how close the model remains to the category of shapes it is supposed to represent
* @param pcaModel model to be evaluated
* @param data test data to verify specificity against
* @param nbSamples number of samples drawn to compute the average
*
* The implementation of this metric is inspired from :
* Styner, Martin A., et al. "Evaluation of 3D correspondence methods for model building." Information processing in medical imaging. Springer Berlin Heidelberg, 2003.
*
* The general idea is as follows :
*
*
* 1 - sample a shape from the mesh model
*
*
* 2- compute the average mesh distance (see [[scalismo.mesh.MeshMetrics]]) of the sample to all elements of the given sequence of meshes and select the minimum distance
*
* These steps are then repeated nbSamples times and the average value is returned.
*
*/
The number 10 indicates that the metric is calculated based on 10 samples. To get a more precise and reliable information, you might have to use there a higher number. See the documentation of the function attached below for more information.
Best, Andreas
/**
* Returns the specificity metric of the Statistical Mesh Model, that is how close the model remains to the category of shapes it is supposed to represent
* @param pcaModel model to be evaluated
* @param data test data to verify specificity against
* @param nbSamples number of samples drawn to compute the average
*
* The implementation of this metric is inspired from :
* Styner, Martin A., et al. "Evaluation of 3D correspondence methods for model building." Information processing in medical imaging. Springer Berlin Heidelberg, 2003.
*
* The general idea is as follows :
*
*
* 1 - sample a shape from the mesh model
*
*
* 2- compute the average mesh distance (see [[scalismo.mesh.MeshMetrics]]) of the sample to all elements of the given sequence of meshes and select the minimum distance
*
* These steps are then repeated nbSamples times and the average value is returned.
*
*/
Elon
Jul 29, 2021, 10:37:05 PM7/29/21
to scalismo
Thank you Marcel for providing the code snippet. However, when evaluating the following line, I receive an IllegalArgumentException:
val dataCollection : DataCollection[_3D, TriangleMesh, EuclideanVector[_3D]] = DataCollection.fromTriangleMesh3DSequence(reference, trainingSet)
Does anyone know why this error is occurring?
Full stack trace:
Exception in thread "main" java.lang.IllegalArgumentException: requirement failed
at scala.Predef$.require(Predef.scala:327)
at scalismo.statisticalmodel.dataset.DataCollection$.differenceFieldToReference(DataCollection.scala:97)
at scalismo.statisticalmodel.dataset.DataCollection$.$anonfun$fromTriangleMesh3DSequence$1(DataCollection.scala:109)
at scala.collection.immutable.ArraySeq.$anonfun$map$1(ArraySeq.scala:71)
at scala.collection.immutable.ArraySeq.$anonfun$map$1$adapted(ArraySeq.scala:71)
at scala.collection.immutable.ArraySeq$.tabulate(ArraySeq.scala:288)
at scala.collection.immutable.ArraySeq$.tabulate(ArraySeq.scala:267)
at scala.collection.ClassTagIterableFactory$AnyIterableDelegate.tabulate(Factory.scala:664)
at scala.collection.immutable.ArraySeq.map(ArraySeq.scala:71)
at scala.collection.immutable.ArraySeq.map(ArraySeq.scala:35)
at scalismo.statisticalmodel.dataset.DataCollection$.fromTriangleMesh3DSequence(DataCollection.scala:108)
at ssm.robustness$.main(robustness.scala:37)
at ssm.robustness.main(robustness.scala)
Marcel Luethi
Aug 2, 2021, 4:47:16 AM8/2/21
to Elon, scalismo
Dear Elon
The typical cause for this message is that your meshes are not in correspondence. You can easily check this by printing out the number of points for each mesh:
val trainingSet : Seq[TriangleMesh[_3D]] = ???Best regards,
for (mesh <- trainingSet) {
println(mesh.pointSet.numberOfPoints)
}
Marcel
To view this discussion on the web visit https://groups.google.com/d/msgid/scalismo/0ad21d78-ef91-4e85-af92-b5aef4153c38n%40googlegroups.com.
Elon
Aug 2, 2021, 2:30:39 PM8/2/21
to scalismo
Hi Marcel,
Thank you, that makes sense. I managed to get everything working.
Cheers,
Elon
Michael Lennon
Aug 13, 2021, 4:02:19 AM8/13/21
to scalismo
Hi Marcel,
I was looking at this previous response by you and have a question about generalisation. In the code snippet you use dataCollection as an input, which I believe contains the meshes that were used to build the SSM. This is in effect the training set of meshes. I understand that you would use the training set on the specificity metric but my interpretation is that generalisation metric should be derived using a test set of meshes against the SSM (as per API doc). So if I want to calculate generality of a model should I use a set of meshes (or a single mesh?) that are independent of the meshes used to train the SSM?
Best regards,
Michael
Marcel Luethi
Aug 16, 2021, 4:18:37 AM8/16/21
to Michael Lennon, scalismo
Hi Michael
Thanks for pointing this out. The correct way to do it is to either have a separate test set or to use Leave-one-out cross validation. Methods for crossvalidation are implemented in Scalismo in the object
scalismo.statisticalmodel.dataset.CrossValidation
Best regards,
Marcel
To view this discussion on the web visit https://groups.google.com/d/msgid/scalismo/1ffea6b9-3d66-4c64-ac2c-20b3564bc543n%40googlegroups.com.
Michael Lennon
Aug 18, 2021, 2:54:29 AM8/18/21
to scalismo
Thank you Marcel,
I am interested in leave-one-out cross validation to compare models. I am unable to work out how to apply this on a simple triangle mesh PDM model created using a dataCollection in Scalismo. I believe I can execute this at a basic level by inputting a data collection and an evaluation function but I am unclear on what the evaluation function would look like so any examples you have would be appreciated. I am also cognisant that I could build different models using one data collection such as a truncated model only using first 5 principal modes. Would your method allow me to compare a metric for full and truncated models?
val referenceMesh: TriangleMesh[_3D] = ???
val trainingMeshes: Seq[TriangleMesh[_3D]] = ???
val dataCollection = DataCollection.fromTriangleMeshSequence(referenceMesh, trainingMeshes)
val pdmTriangleMesh = PointDistributionModel.createUsingPCA(dataCollection)
val truncatedMesh = pdmTriangleMesh.truncate(5)
val generality_full = Crossvalidation.leaveOneOutCrossvalidation(dc, evalFun = ?)
val generality_truncated = Crossvalidation.leaveOneOutCrossvalidation(dc, evalFun = ?)
Best regards,
Michael
Marcel Luethi
Aug 19, 2021, 2:59:42 AM8/19/21
to Michael Lennon, scalismo
Hi Michael,
The following is a basic example of how to use the crossvalidation. You can run it using either a truncated model or a full one. You should see that the truncated model has a much higher generalisation error.
val referenceMesh: TriangleMesh[_3D] = ???
val trainingMeshes: Seq[TriangleMesh[_3D]] = ???
val dataCollection = DataCollection.fromTriangleMesh3DSequence(referenceMesh, trainingMeshes)
// computes whatever metric is appropriate to compare the left out mesh with the model.
// For generalisation it is a projection into the model, followed by the distance
def evalFun(pdm : PointDistributionModel[_3D, TriangleMesh], mesh : TriangleMesh[_3D]) : Double = {
MeshMetrics.avgDistance(pdm.project(mesh), mesh)
}
val generality_full = Crossvalidation.leaveOneOutCrossvalidation(dataCollection, evalFun)
Best regards,
Marcel
To view this discussion on the web visit https://groups.google.com/d/msgid/scalismo/07abcb0a-c1dc-4b3c-90c2-2c7c8937f225n%40googlegroups.com.
Elon
Aug 19, 2021, 4:54:40 PM8/19/21
to scalismo
Hi Marcel,
Thanks for the example code. The function returns a Vector of ArraySeq. Example:
Vector(ArraySeq(0.8292422412339956), ArraySeq(0.8595790418832963), ArraySeq(0.02897680945534543))
I'm assuming we need to take the median of these doubles in order to calculate the generality as described in Styner et al. Is this correct?
Much appreciated,
Elon
Marcel Luethi
Aug 20, 2021, 2:59:55 AM8/20/21
to Elon, scalismo
Hi Elon
Yes, you somehow need to aggregate it. I am not sure though if Styner proposed to use the mean or the median.
Best regards,
Marcel
To view this discussion on the web visit https://groups.google.com/d/msgid/scalismo/7baef859-40b8-45b2-9f50-60fb7813fe07n%40googlegroups.com.
Michael Lennon
Sep 3, 2021, 3:18:10 AM9/3/21
to scalismo
Hi Scalismo community,
Is anyone able to help me with the code to test a truncated model for generality. The precious code from Marcel was helpful for testing the full model as the input is the dataCollection used to build that model plus the evaluation function. However if I truncate the model down to a rank 5 I am unable to work out how to include this in the Crossvalidation. I believe this would be related to the optional "biasModelAndRank" parameter. Any help would be greatly appreciated as I am unable to determine how to do this.
val referenceMesh: TriangleMesh[_3D] = ???
val trainingMeshes: Seq[TriangleMesh[_3D]] = ???
val trainingMeshes: Seq[TriangleMesh[_3D]] = ???
val dataCollection = DataCollection.fromTriangleMesh3DSequence(referenceMesh, trainingMeshes)
val pdmTriangleMesh = PointDistributionModel.createUsingPCA(dataCollection)
val truncatedMesh = pdmTriangleMesh.truncate(5)
// computes whatever metric is appropriate to compare the left out mesh with the model.
// For generalisation it is a projection into the model, followed by the distance
def evalFun(pdm : PointDistributionModel[_3D, TriangleMesh], mesh : TriangleMesh[_3D]) : Double = {
MeshMetrics.avgDistance(pdm.project(mesh), mesh)
}
val generality_full = Crossvalidation.leaveOneOutCrossvalidation(dataCollection, evalFun)
// For generalisation it is a projection into the model, followed by the distance
def evalFun(pdm : PointDistributionModel[_3D, TriangleMesh], mesh : TriangleMesh[_3D]) : Double = {
MeshMetrics.avgDistance(pdm.project(mesh), mesh)
}
val generality_full = Crossvalidation.leaveOneOutCrossvalidation(dataCollection, evalFun)
val generality_truncated = Crossvalidation.leaveOneOutCrossvalidation(dataCollection, evalFun, biasModelAndRank???)
Marcel Luethi
Sep 5, 2021, 4:12:57 AM9/5/21
to Michael Lennon, scalismo
Dear Micheal
Leave one out cross validation works by virtue of the datacollection. It just selects all but one datasets and builts from them a model. The left out one is then used to test. It is not possible to specify the rank of the model directly. But you can do the truncation in the eval fun.
The parameter biasModelAndRank is used to provide an optional model, with which the model built from the data is augmented.
Best regards,
Marcel
To view this discussion on the web visit https://groups.google.com/d/msgid/scalismo/df71bbd5-61e4-47f2-9ebc-2cb02a48ae4cn%40googlegroups.com.
S Arbabi
Dec 15, 2021, 7:16:38 AM12/15/21
to scalismo
Dear Marcel,
In the mentioned paper, Styner plots specificity (vertical axis) for different number of shape modes (horizontal axis).
How can I sample shape model using specific number of shape modes?
Best,
Saeed
Max
Dec 15, 2021, 8:18:32 AM12/15/21
to scalismo
Hi Saeed,
you can easily truncate a model to the first n principal components by calling pdm.truncate(n) (see http://unibas-gravis.github.io/scalismo/latest/api/scalismo/statisticalmodel/PointDistributionModel.html).
Hope this helps.
Best
Max
S Arbabi
Dec 15, 2021, 9:38:17 AM12/15/21
to scalismo
Thank you Max. works good.
This is basically how I calculate specificity:
var curList = List[Double]()
(ssm.rank-1 to 1 by -1).foreach{rank =>
val reduced_ssm = ssm.truncate(rank)
val sample = reduced_ssm.sample
val dists = meshes.map{mesh=>
val dist = MeshMetrics.avgDistance(mesh, sample)
dist
}
curList = min(dists) :: curList
println(rank+" components, result in "+min(dists)+" specificity")
}
(ssm.rank-1 to 1 by -1).foreach{rank =>
val reduced_ssm = ssm.truncate(rank)
val sample = reduced_ssm.sample
val dists = meshes.map{mesh=>
val dist = MeshMetrics.avgDistance(mesh, sample)
dist
}
curList = min(dists) :: curList
println(rank+" components, result in "+min(dists)+" specificity")
}
is it looking correct? I want to have charts like Styner et al for specificity.
I see that the changes in specificity are very small by increasing the number of shape modes. is there something I'm missing?
Best,
Saeed
Max
Dec 15, 2021, 10:34:30 AM12/15/21
to scalismo
Your code looks fine to me. However, I would suggest sampling more than one time as this improves reliability (since you average over a lot of samples). Regarding your second question, I would say it is normal for specificity to converge after a few principal components (this is at least what I observed so far).
Best
Max
S Arbabi
Dec 16, 2021, 4:04:33 AM12/16/21
to scalismo
Hi Michael,
Were you able to do the generalization with different ranks?
Could you share a bit here?
Could you share a bit here?
Best
Michael Lennon
Dec 18, 2021, 1:40:31 AM12/18/21
to S Arbabi, scalismo
Hi Saeed,
My code for calculating generality for a truncated model is below. Simply add the truncate function to the pdm parameter in evalFun. In the example below I set rank to 3. I also have some code to take the output which is a Vector of ArraySeq and calculate the average distance value from the vector of cross validation results.
def evalFun(pdm : PointDistributionModel[_3D, TriangleMesh], mesh : TriangleMesh[_3D]) : Double = {
MeshMetrics.avgDistance(pdm.truncate(3).project(mesh), mesh)
}
val generality = Crossvalidation.leaveOneOutCrossvalidation(dc, evalFun)
val generality_vector = for (i <- 0 until generality.length) yield (generality(i).head)
val avg_generality = generality_vector.sum / generality_vector.length
To view this discussion on the web visit https://groups.google.com/d/msgid/scalismo/d4103e00-7d18-4b55-be56-9eeacf5dcae3n%40googlegroups.com.
Bereket Workie Anteneh
Dec 21, 2021, 6:22:47 AM12/21/21
to Michael Lennon, S Arbabi, scalismo
Hi Michael,
I want to do generalization test based on truncated model that you provided,
But what does the truncate (- ) the number 3 indicate? or Is there any standard to use which truncated number?
With regards,
Bereket
You received this message because you are subscribed to a topic in the Google Groups "scalismo" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/scalismo/SR5TKFxJc1Y/unsubscribe.
To unsubscribe from this group and all its topics, send an email to scalismo+u...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/scalismo/CAPvL3iE2XaCUJ7ijE__HfHH1JMgCkwYEThcZAzM%3DPhWOioTEpw%40mail.gmail.com.
Michael Lennon
Dec 22, 2021, 8:22:46 PM12/22/21
to Bereket Workie Anteneh, S Arbabi, scalismo
Hi Bereket,

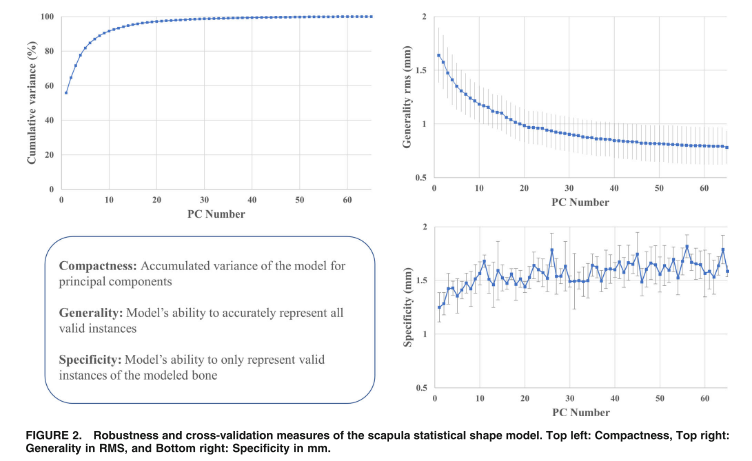
I should point out that I am not part of the scalismo team and not an expert on the maths of GPMM's. But I will attempt to answer your question to support this community. The number 3 in my example is the rank of the model that I want to evaluate. In my case I had 59 training shapes so that is the maximum rank for my model. I recommend you review Marcel's paper "Gaussian Process Morphable Models" to understand parameterising the model using Principal Component Analysis (see extract below). Without truncating, my model would be very complex and would rely on 59 parameters (n = 59). If I want to truncate the high rank model I have down to only the first 3 components (n = 3) I use the truncate method on my model. I believe this characteristic of models is referred to as Compactness and I recommend you do further research on that idea. If I wanted a very compact model, described only with 3 principal components (n = 3) then I would want to evaluate the generality and specificity of that low rank representation and compare to less compact models with say a rank of 10. I can also recommend a paper by Salhi et al called "Statistical Shape Modeling Approach to Predict Missing Scapular Bone". I have pasted an extract from that below where the authors have plotted compactness, generality and specificity against the number of principal components (rank). I hope this helps.


Message has been deleted
Marcel Luethi
Dec 29, 2021, 1:56:48 PM12/29/21
to Bereket Workie Anteneh, scalismo
Dear all,
Is it possible that we close this thread and start a new one?
The thread has become so complicated that gmail is not helpful anymore and I don't know , which question has been answered and what is still open.
Best,
Marcel
On Thu, Jul 15, 2021 at 2:25 PM Bereket Workie Anteneh <join...@gmail.com> wrote:
Thank you Marcel for sharing what I want,But still now I have a question specially in the code line 2 that is "val trainingData : Seq[TriangleMesh[_3D]] = ???"The path of my training data was at "C:\\Users\\user\\Documents\\minimal-scalismo-seed-master\\data\\2Projecetd Meshes\\"When I try to declare the path as shown belowval trainingData : Seq[TriangleMesh[_3D]] = MeshIO.readMesh(new File("C:\\Users\\user\\Documents\\minimal-scalismo-seed-master\\data\\2Projecetd Meshes\\")).getIt says an error, it says required: "Seq[TriangleMesh[_3D]] found: TriangleMesh [_3D]".I tried a lot to fix it but it challenges me. So what would be the correct path declaration based on the training data directory that I mentioned above?With regards,
To view this discussion on the web visit https://groups.google.com/d/msgid/scalismo/d1bb0671-efc3-4814-8326-bc491e7a2bc0n%40googlegroups.com.
Bereket Workie Anteneh
Dec 30, 2021, 6:47:33 AM12/30/21
to Marcel Luethi, scalismo
Dear Scalismo community,
I want to measure the angle of inclination of the neck of femur, and diameter of the neck and diameter of femoral head. I tried a lot but it challenges me.
So, is there a possible solution with Scalismo, especially a snippet of code that can measure the listed issues?
With regards,
Bereket
Reply all
Reply to author
Forward
0 new messages
