QTL analysis with MAGIC population
68 views
Skip to first unread message
Ramesh Bhat
Aug 14, 2020, 6:46:46 AM8/14/20
to R/qtl2 discussion
Dear Dr. Karl,
I have a peanut MAGIC population of 500 lines derived from eight parents with the cross type of risib8.
Is this YAML file okay for risib8?
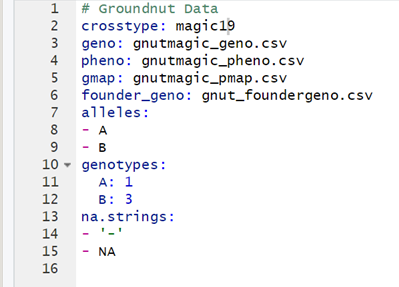
Also, I am getting the following error, kindly help
Kind regards,
Ramesh S. Bhat
Professor and HeadDepartment of Biotechnology
University of Agricultural Sciences, Dharwad
PIN: 580 005, Dharwad, Karnataka, India
Ph: +91-836-2214457, Mobile: +91-9945667300
Karl Broman
Aug 14, 2020, 8:22:47 AM8/14/20
to R/qtl2 discussion
"crosstype: magic19" is not right, as you said the cross type was risib8.
Consider the CC example at https://github.com/rqtl/qtl2data/tree/master/CC
karl
Ramesh Bhat
Aug 14, 2020, 9:56:28 AM8/14/20
to R/qtl2 discussion
--
You received this message because you are subscribed to the Google Groups "R/qtl2 discussion" group.
To unsubscribe from this group and stop receiving emails from it, send an email to rqtl2-disc+...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/rqtl2-disc/5648daae-264b-48fe-9557-e181e2fe23acn%40googlegroups.com.
Karl Broman
Aug 14, 2020, 10:45:47 AM8/14/20
to R/qtl2 discussion
The files are available at the link I gave you, including as a zip file, cc.zip.
karl
Ramesh Bhat
Aug 14, 2020, 12:18:28 PM8/14/20
to R/qtl2 discussion
Thanks. I could download that zip file.
Got the following error
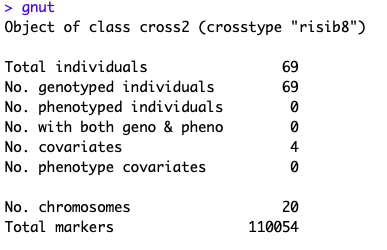
What shall I do?
To view this discussion on the web visit https://groups.google.com/d/msgid/rqtl2-disc/562e05d8-6ef0-4a9c-b18f-72b5331e540en%40googlegroups.com.
Ramesh Bhat
Aug 14, 2020, 12:40:32 PM8/14/20
to R/qtl2 discussion
Not able to find any cc_pheno.csv in cc.zip. Is this the reason to get the error?
To view this discussion on the web visit https://groups.google.com/d/msgid/rqtl2-disc/562e05d8-6ef0-4a9c-b18f-72b5331e540en%40googlegroups.com.
Karl Broman
Aug 14, 2020, 4:16:10 PM8/14/20
to R/qtl2 discussion
These data include no phenotypes. I thought they might be useful to you as an example, to structure your own data.
karl
Ramesh Bhat
Aug 14, 2020, 9:35:53 PM8/14/20
to R/qtl2 discussion
Thanks Dr. Karl.
We have phenotyped the peanut MAGIC population over four seasons. SNP data will be available in about 1-2 months.
Meanwhile, can I get acquainted with the scripts using Arabidopsis 19-way example?
Later, I would like to analyze our MAGIC population for
1. QTL and marker for the traits
2. Effects (additive) for the QTL or the marker
3. PVE for the QTL/marker
4. Identifying the parent with the allele having higher mean
Kind regards,
To view this discussion on the web visit https://groups.google.com/d/msgid/rqtl2-disc/6ecfc571-cb11-45dc-94b4-1e9e11cddc97n%40googlegroups.com.
Ramesh Bhat
Aug 16, 2020, 11:45:43 PM8/16/20
to R/qtl2 discussion
Dear Dr. Karl,
I am working with ArabMAGIC. The following function creates the zip file on my Macbook, but not on my Windows desktop.
zip_datafiles(control_file, "gnutmagic.zip")
Does this require any change?
I got the effects with the following function
c2eff <- scan1coef(pr[,"2"], gnut$pheno[,"height"])
What formula does it consider to calculate the effets here? What do they mean?
Kind regards,
On Sat, Aug 15, 2020 at 1:46 AM Karl Broman <kbr...@gmail.com> wrote:
To view this discussion on the web visit https://groups.google.com/d/msgid/rqtl2-disc/6ecfc571-cb11-45dc-94b4-1e9e11cddc97n%40googlegroups.com.
Karl Broman
Aug 18, 2020, 6:41:49 PM8/18/20
to R/qtl2 discussion
I can't tell what is going wrong with zip_datafiles() without further information, such as the exact error message you're getting.
scan1coef() estimates effects via linear regression.
karl
Ramesh Bhat
Aug 18, 2020, 11:51:41 PM8/18/20
to R/qtl2 discussion
Unfortunately, it does not show any error, but the zip file is not created.
To view this discussion on the web visit https://groups.google.com/d/msgid/rqtl2-disc/d60bb9ba-3e50-461c-9d83-acb7d3bac21fn%40googlegroups.com.
Ramesh Bhat
Aug 20, 2020, 7:16:16 AM8/20/20
to R/qtl2 discussion
Dear Dr. Karl,
Can we get the effects and gene annotations for the SNPs using R/qtl2 especially for peanut?
Kind regards,
To view this discussion on the web visit https://groups.google.com/d/msgid/rqtl2-disc/d60bb9ba-3e50-461c-9d83-acb7d3bac21fn%40googlegroups.com.
Reply all
Reply to author
Forward
0 new messages
