biparental population QTL analysis
29 views
Skip to first unread message
venkat rami reddy
Sep 30, 2021, 8:57:55 AM9/30/21
to rqtl...@googlegroups.com
Hi Karl,
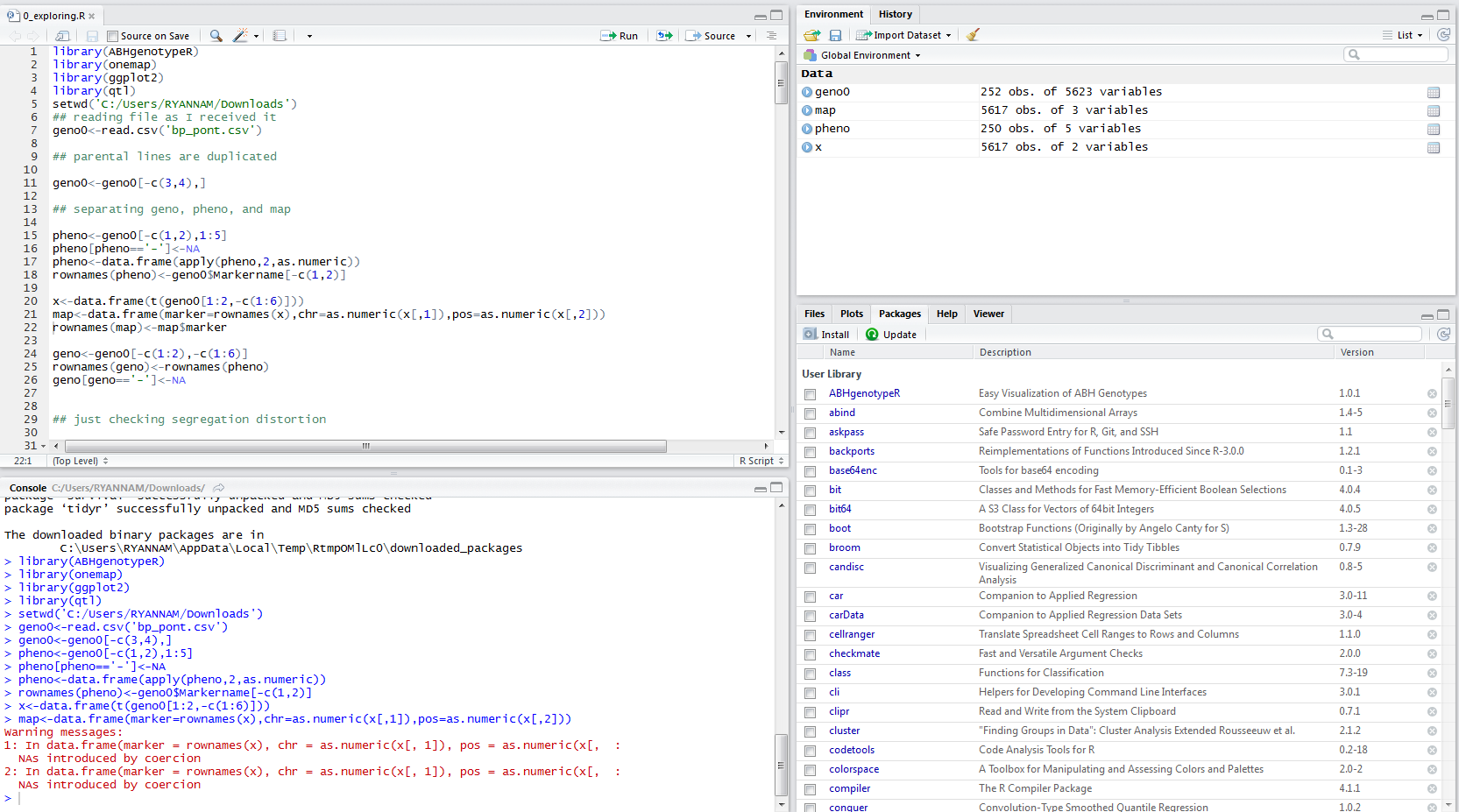
I have an error while running the script to separate genotype, phenotype, and map. I didn't get the point as the missing datas are introduced as NAs. i have tried to run the following script to perform segregation script but i have an error while f<-which(seg[,3]>10)
The code and the warning is as below. can you please explain me and help me out with the error
I have also attached the screenshot and the script for your reference......
geno0<-read.csv('bp_pont.csv')
> geno0<-geno0[-c(3,4),]
> pheno<-geno0[-c(1,2),1:5]
> pheno[pheno=='-']<-NA
> pheno<-data.frame(apply(pheno,2,as.numeric))
> rownames(pheno)<-geno0$Markername[-c(1,2)]
>
> x<-data.frame(t(geno0[1:2,-c(1:6)]))
> map<-data.frame(marker=rownames(x),chr=as.numeric(x[,1]),pos=as.numeric(x[,2]))
Warning messages:
1: In data.frame(marker = rownames(x), chr = as.numeric(x[, 1]), pos = as.numeric(x[, :
NAs introduced by coercion
2: In data.frame(marker = rownames(x), chr = as.numeric(x[, 1]), pos = as.numeric(x[, :
NAs introduced by coercion
> rownames(map)<-map$marker
>
> geno<-geno0[-c(1:2),-c(1:6)]
> rownames(geno)<-rownames(pheno)
> geno[geno=='-']<-NA
> seg<-t(apply(geno,2,function(x){
+ w<-c(0,0,0)
+ names(w)<-c('A','B','H')
+ y<-table(x)
+ w[names(y)]<-y
+ return(w)
+ }))
>
>
> f<-which(seg[,3]>10)
Error in which(seg[, 3] > 10) :
'list' object cannot be coerced to type 'double'
> geno<-geno[,-f]
Error in `[.data.frame`(geno, , -f) : object 'f' not found
> map<-map[-f,]
Error in `[.data.frame`(map, -f, ) : object 'f' not found
> geno0<-geno0[-c(3,4),]
> pheno<-geno0[-c(1,2),1:5]
> pheno[pheno=='-']<-NA
> pheno<-data.frame(apply(pheno,2,as.numeric))
> rownames(pheno)<-geno0$Markername[-c(1,2)]
>
> x<-data.frame(t(geno0[1:2,-c(1:6)]))
> map<-data.frame(marker=rownames(x),chr=as.numeric(x[,1]),pos=as.numeric(x[,2]))
Warning messages:
1: In data.frame(marker = rownames(x), chr = as.numeric(x[, 1]), pos = as.numeric(x[, :
NAs introduced by coercion
2: In data.frame(marker = rownames(x), chr = as.numeric(x[, 1]), pos = as.numeric(x[, :
NAs introduced by coercion
> rownames(map)<-map$marker
>
> geno<-geno0[-c(1:2),-c(1:6)]
> rownames(geno)<-rownames(pheno)
> geno[geno=='-']<-NA
> seg<-t(apply(geno,2,function(x){
+ w<-c(0,0,0)
+ names(w)<-c('A','B','H')
+ y<-table(x)
+ w[names(y)]<-y
+ return(w)
+ }))
>
>
> f<-which(seg[,3]>10)
Error in which(seg[, 3] > 10) :
'list' object cannot be coerced to type 'double'
> geno<-geno[,-f]
Error in `[.data.frame`(geno, , -f) : object 'f' not found
> map<-map[-f,]
Error in `[.data.frame`(map, -f, ) : object 'f' not found

thanks and regards
Rami
Karl Broman
Sep 30, 2021, 9:14:35 AM9/30/21
to R/qtl discussion
I would suggest talking to the person that wrote the script.
It's hard to debug such code without access to the data, and this list is focused on R/qtl, and so this script doesn't seem within the group's scope.
karl
Reply all
Reply to author
Forward
0 new messages
