New Studies Enable a Clearer View Inside Cells
rael-science
New Studies Enable a Clearer View Inside Cells
Armed
with improved imaging techniques and supercomputers, researchers are
generating detailed three-dimensional images of cellular structures that
anyone can explore.
Andrew Chapman
Nov 4, 2021
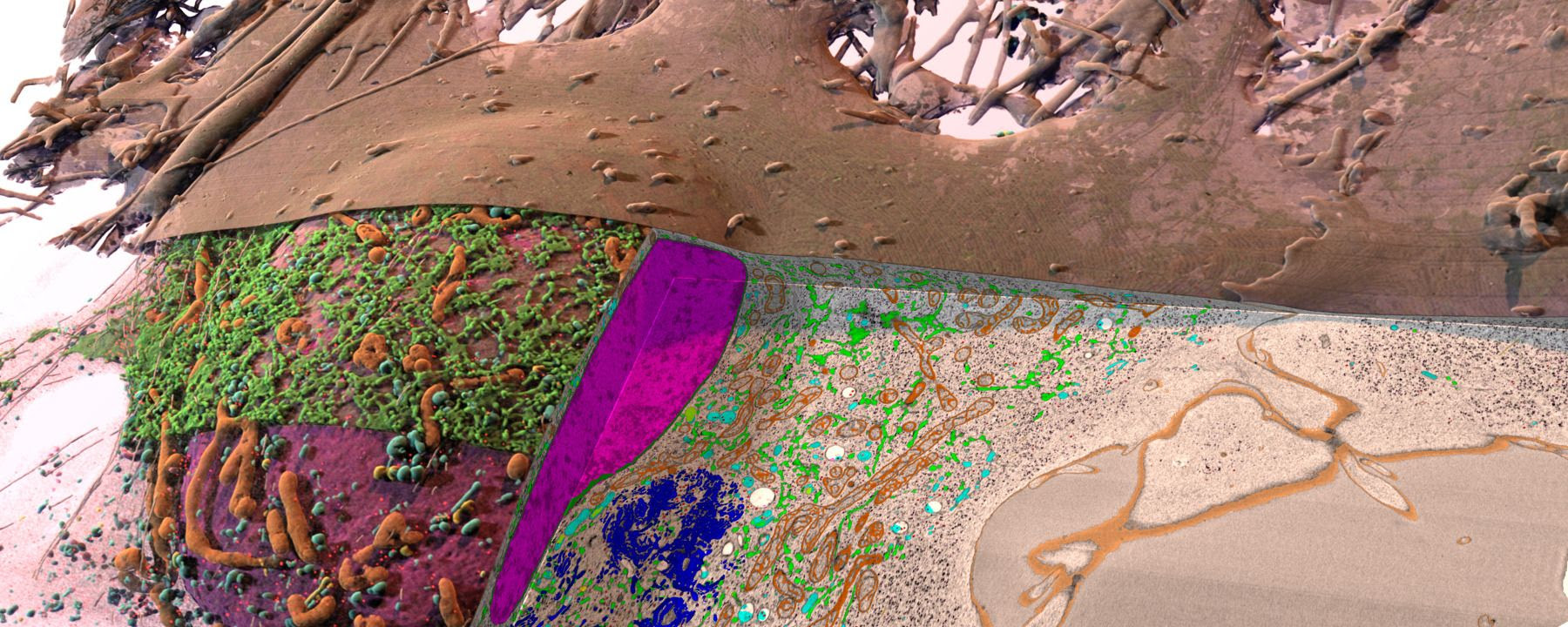
ABOVE: 3D rendering of a HeLa cell: plasma membrane (brown), ER (green),
mitochondria (orange), nucleus (purple), Golgi (blue), endosomes (cyan),
vesicles (red), lysosomes (yellow), lipid droplets (pink), microtubules
(dark sticks), and ribosomes (pink haze) COURTESY OF STEPHAN SAALFELD
To
fully understand how cells work, scientists need to know how their
moving parts relate to one another in space and time. However, because
of their size and the amount of data involved, visualizing cellular
structures in three dimensions has proven difficult. Now, in a trio of
new studies, two teams of molecular scientists have aimed to make it
easy for everyone to see inside cells. By incorporating painstakingly
collected experimental data and partnering with computational
biologists, they are bringing 3D visualizations of organelles and
chromosomes into sharper focus.
The researchers are also making
their 3D data, published in separate studies in early October, freely
available for anyone to explore in order to allow researchers around the
globe to probe their own questions about how cellular form impacts
function. As Karissa Sanbonmatsu, a structural biologist at Las Alamos
National Laboratory and coauthor on one of the papers, puts it: “We’re
trying to do Google Earth for chromosomes.”
Using the techniques from these three papers, computational biologist Robert Murphy of Carnegie Mellon University, who wasn’t involved in the research,
says scientists might compare different cell types—for example, a cancer
cell and a healthy cell—to start to understand what role the
organization of cellular structures plays in physiology and disease.
“That’s one of the very first things you would want to do.”
Improving organelle clarity, with a little help from AI
For
the past decade, Howard Hughes Medical Institute (HHMI) scientist Shan
Xu has worked to adapt focused ion beam scanning electron microscopy
(FIB-SEM)—a microscopy technique originally developed for material
science applications—to biological research. FIB-SEM works by taking an
SEM image of objects embedded in resin, then shaving a tiny sliver of
the sample using the ion beam and taking another picture. By repeating
this process over and over, scientists can stack all the images to
create a 3D rendering. But the machines need to shut down after 3 to 5
days to recharge the ion beam, and when they start up again, the amount
of material the beam shaves off and the resolution of the image aren’t
as accurate. To reduce the inaccuracy after each shutdown and achieve
the clarity needed to map the insides of cells, Xu added more stable ion
beam hardware that allowed a higher current, troubleshot the control of
the beam, and sped up the imaging of the SEM. Xu constructs each of the
machines himself. “I build them one by one, so [they’re] like my kids,”
he says.
Those tweaks, detailed in one of his team’s two October Nature papers, improved the 3D resolution to 4 nm from 8 nm, which often meant
the difference between organelles looking like clear, detailed
structures and fuzzy clouds.
But the resulting volume of data
meant that the researchers needed a faster way to identify and map out
the organelles inside the cells: enter machine learning. For two years,
two people worked full-time to manually identify organelles and outline
their boundaries in 3D images from FIB-SEM. Then, Larissa Heinrich, a
computer scientist at HHMI Janelia Research Campus, used those
annotations to train a neural network to map structures within the
cells, as reported in the team’s second Nature paper.
Heinrich
says that the network uses the manually annotated images to learn
rules, “trying to adjust them in a way that the output it produces is
the same as what the humans did.” The network doesn’t just look at each
pixel and make a call about whether it’s part of an organelle; it
examines the pixels around it to determine whether the call is logical.
The scientists estimate it would have taken one person 60 years to
manually identify the same number of organelles the algorithms can map
in a few hours.
Murphy says the results of the two studies show “a
critical instrumentation advance,” adding that “the use of that
technology to produce the large-scale data collections that they’ve done
is important.” He says that the machine learning work is vital, but
notes that the algorithms still can’t identify every type of organelle
with a high level of accuracy. The discrepancy in identification is
often related to how abundant the organelles are in the human-labeled
training sets. For example, centrosomes are rare in each cell, so the AI
doesn’t have as many chances to learn what they look like from the
training sets. Heinrich says that more training sets and algorithms will
help improve the mapping accuracy for all organelles and structures
present inside cells.
Still, with the high-resolution images and
the human- and AI-identified organelles, the researchers were able to
build open-access 3D atlases of several cells and tissues, including commonly used HeLa cells,
immune T-cells attacking ovarian cancer cells, and pancreatic
beta-cells.
Adding the fourth dimension
In the third study, published in PNAS, a separate research group used computational approaches to infer the 3D structure of chromosomes.
In
the past, to infer 3D DNA structures, scientists have first used a
technique called Hi-C to determine the 2D interactions between sections
of DNA. Hi-C involves physically cross-linking interacting stretches of
DNA and then fusing them together. That way, all the interacting
stretches of DNA, even if they are far apart on the chromosome, will
appear side by side in the sequencing data. These 2D data would then be
used to build 3D models, but this required a lot of assumptions about
what would happen based on the DNA sequence and proteins that hold loops
of DNA together. The researchers behind the new study didn’t want to
make that many assumptions. “All we wanted to do is simply make a
structure that follows the rules from experiments,” says the study’s
first author Anna Lappala, a polymer physicist at Massachusetts General
Hospital.
So the researchers incorporated experimental 2D
interaction data, simulated physical forces, and Newton’s equations of
motion to predict the 3D structure of the X chromosome. They didn’t stop
with 3D, however. They repeated the process at different time points
during a process called X chromosome inactivation (XCI), thus adding the
fourth dimension to their analysis. The high-resolution modeling, which
required analysis of enormous datasets, was made possible by using
supercomputers at Los Alamos National Laboratory.
When
two X chromosomes are present in a cell, most of the genes on one X are
deactivated through XCI to prevent developmental abnormalities. This
silencing is initiated when one X expresses a noncoding RNA called Xist that coats the chromosome.
The
results of the structural modeling show that as the X chromosome
undergoes XCI, it forms a dense core with a looser surface. Several
genes known to escape XCI are located at the surface, which the researchers suggest might allow
better access for the molecular machinery that expresses genes. “What’s
really ground-breaking about this paper is that we figured out a way to
visualize the 3D structure of the X chromosome based on a decidedly
two-dimensional map that one gets from Hi-C datasets,” says Jeannie Lee,
a molecular biologist at Harvard Medical School and coauthor on the
paper. The authors were also able to track the spread of Xist RNA on the chromosome over time, which they show in a video.
Sanbonmatsu,
also an author on the paper, says that their computational approach to
modeling 3D structure could also be applied to the rest of the
chromosomes in the genome.
Computing the future of biology
“What I think is most significant about this kind of work [in the PNAS paper]
is the attempt to explain something very complex, such as the structure
of the genome and especially dynamic processes in the genome, based on
the fundamental first principles of physics,” says biomedical engineer
Vadim Backman of Northwestern University who wasn’t involved in any of
the three studies.
“Across all three papers, an important point
to make is how critical computational analysis and modeling is for this
area,” adds Murphy. In the three new papers, powerful computers allowed
the scientists to take huge experimental datasets and learn something
new about organelles, chromosomes, and other structures without years
and years of manual work.
To speed up the analysis of data and
progression of 3D cellular biology research even more, the authors of
all the papers are committed to open access to data. “It’s much better
to let the world see what we have invested in,” Xu says. Xu has patented
the new microscope technology, but it is free for universities and
nonprofits to use, and the atlas of 3D cell data is freely available to
explore. Sanbonmatsu wants to eventually enable biologists to look at
the 3D structure of whichever chromosome or gene they are interested in
using a “point and click on a browser.”
“We’re trying to democratize this whole process,” she says.
