Three way interactions for Poisson-Multinomial transformed models
78 views
Skip to first unread message
Adam Howes
Nov 24, 2021, 8:27:06 AM11/24/21
to R-inla discussion group
Hi R-INLA mailing list,
I've been working on a spatio-temporal multinomial model in R-INLA using the Poisson-Multinomial transformation. This involves separating data of the form y = (y_1, ..., y_K) into a separate row in the model for each of the K categories.
I've added the two-way interaction random effects to the model, using the R-INLA group option to define Gaussian Kronecker Markov random fields, such as {space x category} and {time x category}.
Now what I'd like to do is define three-way interaction random effects to represent {space x time x category}, including:
I've been working on a spatio-temporal multinomial model in R-INLA using the Poisson-Multinomial transformation. This involves separating data of the form y = (y_1, ..., y_K) into a separate row in the model for each of the K categories.
I've added the two-way interaction random effects to the model, using the R-INLA group option to define Gaussian Kronecker Markov random fields, such as {space x category} and {time x category}.
Now what I'd like to do is define three-way interaction random effects to represent {space x time x category}, including:
a) IID x IID x IID,
b) Besag x IID x IID,
c) IID x AR1 x IID,,
d) Besag x AR1 x IID.
I have found that it is possible to do this (without using the rgeneric) as follows:
a) f(area_sur_idx, model = "iid", group = cat_idx, control.group = list(model = "iid"), constr = TRUE, ...),
b) f(area_sur_idx, model = "besag", graph = interaction_adjM_6x, scale.model = TRUE, group = cat_idx, control.group = list(model = "iid"), constr = TRUE, ...),
c) f(sur_idx, model = "ar1", group = area_cat_idx, control.group = list(model = "iid"), constr = TRUE, ...),
d) f(area_cat_idx, model = "besag", graph = interaction_adjM_9x, scale.model = TRUE, group = sur_idx, control.group = list(model = "ar1", ...), constr = TRUE, ...),
where "area" are space, "sur" are survey (time) and "cat" are category indicators. interaction_adjM_nx are adjacency matrices which are larger versions of the area adjacency matrix created by copying the adjacency matrix along the diagonal a number of times.
These work alright I think, but the problem I am now running into is how to get the constraints to be correct. I believe that a) and b) above have the correct constraints, whereby within each category there is a sum-to-zero. However, because of the way that I have written c) and d) the group option is no longer set to just cat_idx and the constraints are different.
My supervisor Jeff Eaton suggested that I try defining separate f terms for each category and using the copy option to make sure the precision parameter is the same. The other option (a bit of a worst case) is to write custom rgeneric models for IID x AR1 and Besag x AR1.
Thanks for making it this far! There are some more details about what I have tried at this issue on GitHub (this is probably the most helpful comment). If anyone is able to help out, my questions are:
* Is what I have done reasonable? Or am I overcomplicating something which can be done easier another way.
* Is the copy option a good alternative to try?
* Has anyone already written a rgeneric for IID x AR1 or Besag x AR1?
Best wishes,
Adam Howes
I have found that it is possible to do this (without using the rgeneric) as follows:
a) f(area_sur_idx, model = "iid", group = cat_idx, control.group = list(model = "iid"), constr = TRUE, ...),
b) f(area_sur_idx, model = "besag", graph = interaction_adjM_6x, scale.model = TRUE, group = cat_idx, control.group = list(model = "iid"), constr = TRUE, ...),
c) f(sur_idx, model = "ar1", group = area_cat_idx, control.group = list(model = "iid"), constr = TRUE, ...),
d) f(area_cat_idx, model = "besag", graph = interaction_adjM_9x, scale.model = TRUE, group = sur_idx, control.group = list(model = "ar1", ...), constr = TRUE, ...),
where "area" are space, "sur" are survey (time) and "cat" are category indicators. interaction_adjM_nx are adjacency matrices which are larger versions of the area adjacency matrix created by copying the adjacency matrix along the diagonal a number of times.
These work alright I think, but the problem I am now running into is how to get the constraints to be correct. I believe that a) and b) above have the correct constraints, whereby within each category there is a sum-to-zero. However, because of the way that I have written c) and d) the group option is no longer set to just cat_idx and the constraints are different.
My supervisor Jeff Eaton suggested that I try defining separate f terms for each category and using the copy option to make sure the precision parameter is the same. The other option (a bit of a worst case) is to write custom rgeneric models for IID x AR1 and Besag x AR1.
Thanks for making it this far! There are some more details about what I have tried at this issue on GitHub (this is probably the most helpful comment). If anyone is able to help out, my questions are:
* Is what I have done reasonable? Or am I overcomplicating something which can be done easier another way.
* Is the copy option a good alternative to try?
* Has anyone already written a rgeneric for IID x AR1 or Besag x AR1?
Best wishes,
Adam Howes
INLA help
Nov 24, 2021, 10:56:21 AM11/24/21
to Adam Howes, R-inla discussion group
Its possible to do
Model x IID x AR1
With f(…, replicate=…, group=…, etc…)
But the constraints will ONLY be on ‘Model’, and will be replicated over ‘IID x AR1’
If you need other constraints, then using ‘rgeneric’ is the way to go, and its less involved than you might think.
What constraints do you need? An alternative is to use a non-intrinsic model where constraints are not needed, or if confounding is a issue, you can extract posteriors for contrasts.
There are good (non-exported) functions in the package for dealing with the ‘besag’ model, to deal with scaling, and automatic correction for single-site regions and disconnected regions.
--
You received this message because you are subscribed to the Google Groups "R-inla discussion group" group.
To unsubscribe from this group and stop receiving emails from it, send an email to r-inla-discussion...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/r-inla-discussion-group/bb4793b4-ffa0-4c5e-b38c-ade328aa8470n%40googlegroups.com.
Adam Howes
Nov 25, 2021, 6:05:43 AM11/25/21
to R-inla discussion group
Thank you for your help!
The goal is to interpret the precision parameter of these random effects in a variance decomposition like this (adding Area x Survey x Category):
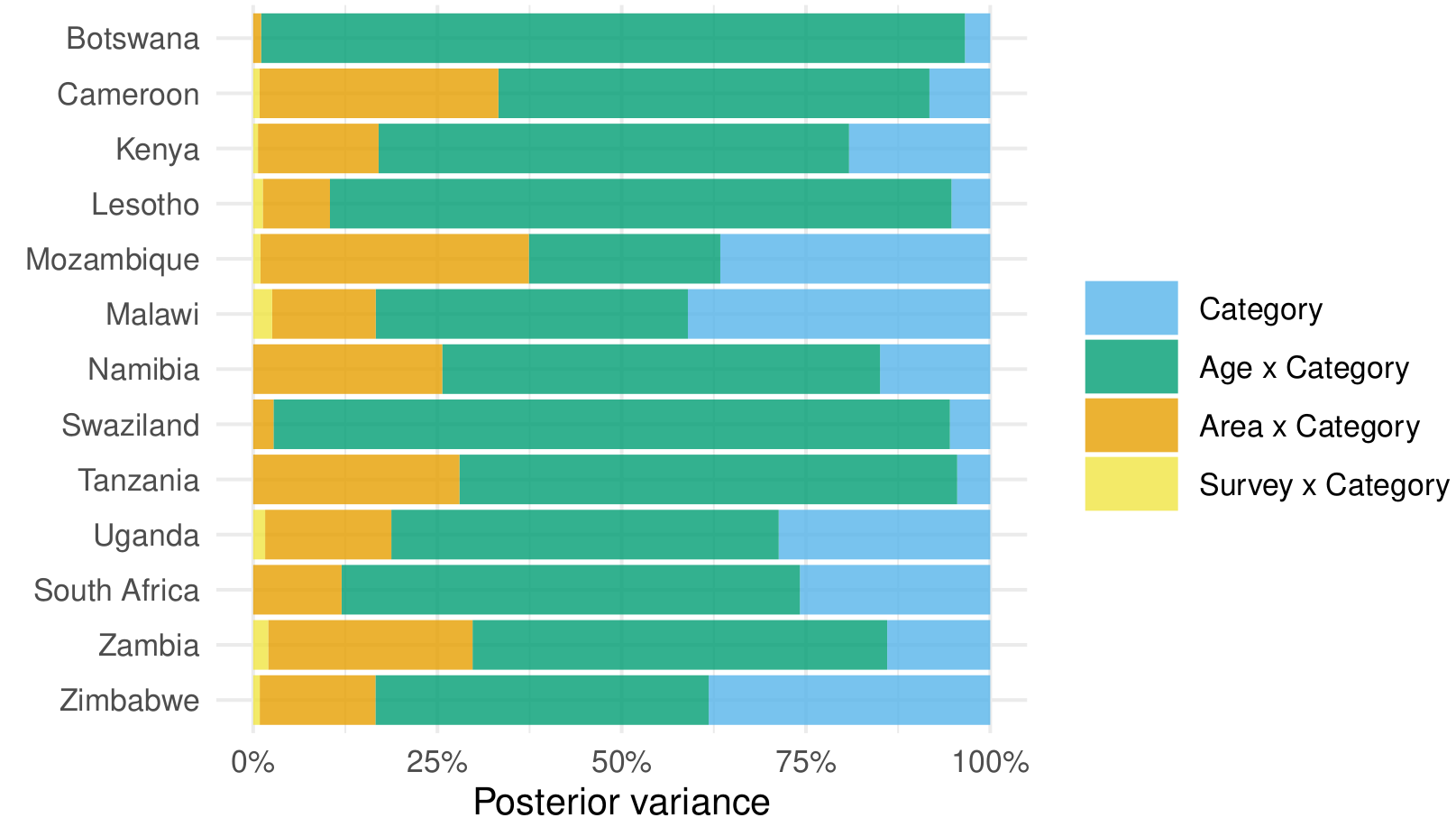
The most prominent thing that comes to mind is that I'd like a sum-to-zero so that the effects do not alter the overall category probabilities.
Let the three-way interaction random effects be \delta_{itk} where i is space, t is time and k is category, then this would be:
A) sum_{it} \delta_{itk} = 0 \forall k = 1, ..., K.
That said, I think I just have the category constraints in my mind because this is what I was thinking about when implementing the two-way interactions above.
To stop the three-way interactions altering the overall level of the space or time, perhaps (?) I also want:
B) sum_{ik} \delta_{itk} = 0 \forall t,
C) sum_{tk} \delta_{itk} = 0 \forall i .
The goal is to interpret the precision parameter of these random effects in a variance decomposition like this (adding Area x Survey x Category):

The most prominent thing that comes to mind is that I'd like a sum-to-zero so that the effects do not alter the overall category probabilities.
Let the three-way interaction random effects be \delta_{itk} where i is space, t is time and k is category, then this would be:
A) sum_{it} \delta_{itk} = 0 \forall k = 1, ..., K.
That said, I think I just have the category constraints in my mind because this is what I was thinking about when implementing the two-way interactions above.
To stop the three-way interactions altering the overall level of the space or time, perhaps (?) I also want:
B) sum_{ik} \delta_{itk} = 0 \forall t,
C) sum_{tk} \delta_{itk} = 0 \forall i .
I think that the replicate option you describe above would be getting something like:
D) sum_{i} \delta_{itk} = 0 \forall k, t
And that D implies A and B, but it wouldn't get C? If this is this case it's somewhat tempting to accept some misspecification in the model and see if it plays a huge role in the posterior (i.e. checking the size of sums over tk).
Do you have a reference where I can read more on the "extract posteriors for contrasts" idea? I've heard it mentioned before but not familiar.
> An alternative is to use a non-intrinsic model where constraints are not needed.
My concern is related to e.g. {Area x Survey x Category} interactions stealing the effects of e.g. {Area x Category} random effects. I suppose this is what's called confounding in this context. Another way to say might be that it's creating an unidentified model whereby variance can be explained with either {Area x Survey x Category} or {Area x Category} equally. That said, I haven't been thinking very carefully about how the more peculiar requirements of the ICAR sum-to-zero constraints might play into this.
And that D implies A and B, but it wouldn't get C? If this is this case it's somewhat tempting to accept some misspecification in the model and see if it plays a huge role in the posterior (i.e. checking the size of sums over tk).
Do you have a reference where I can read more on the "extract posteriors for contrasts" idea? I've heard it mentioned before but not familiar.
> An alternative is to use a non-intrinsic model where constraints are not needed.
My concern is related to e.g. {Area x Survey x Category} interactions stealing the effects of e.g. {Area x Category} random effects. I suppose this is what's called confounding in this context. Another way to say might be that it's creating an unidentified model whereby variance can be explained with either {Area x Survey x Category} or {Area x Category} equally. That said, I haven't been thinking very carefully about how the more peculiar requirements of the ICAR sum-to-zero constraints might play into this.
Esmail Abdul Fattah
Nov 25, 2021, 1:52:42 PM11/25/21
to R-inla discussion group
Hello Adam,
Here is a way how to deal with constraints.
Given any model combination: for example Q = model x model or Q = model x model x model
1. Use rgeneric
2. Set constr=FALSE
3. Use extraconstr = CON, such that CON <- list(A = t(eigen(Q)$vectors[m-r+1,m]), e = rep(0, r)), m is the dimension of Q and R = m - r is the rank of Q or the dim of eigen(Q)$vectors[m-r+1,m]) is m by r (the number of constraints).
Best Regards,
Esmail
Reply all
Reply to author
Forward
0 new messages
