calculate posterior group-level means
37 views
Skip to first unread message
Ramona
Nov 23, 2021, 1:01:16 PM11/23/21
to R-inla discussion group
Hi all,
I am completely new to R INLA and currently trying to calculate posterior group-level means and CrI's of the posterior predictive distribution.
Is there a function like fitted() in brms? (https://rdrr.io/cran/brms/man/fitted.brmsfit.html)And if nut, what is the best way to calculate it?
Thanks a lot!!!
Helpdesk
Nov 23, 2021, 11:24:25 PM11/23/21
to Ramona, R-inla discussion group
Hi
I would check out
https://www.r-inla.org/learnmore/books
and especially the book by Virgilo Gomez-Rubio which is also avaialble
online.
Best
H
> You received this message because you are subscribed to the Google
> Groups "R-inla discussion group" group.
> To unsubscribe from this group and stop receiving emails from it, send
> an email to r-inla-discussion...@googlegroups.com.
> To view this discussion on the web, visit
> https://groups.google.com/d/msgid/r-inla-discussion-group/5defa1ff-aad5-4562-a282-d12408c2efb1n%40googlegroups.com
> .
--
Håvard Rue
he...@r-inla.org
Ramona
Nov 26, 2021, 1:09:52 PM11/26/21
to R-inla discussion group
Dear Havard Rue,
thank you for this advice! Before buying books, I would like to be sure that there is a solution for my problem to be found.
To be more concrete, here my model structure:
formula <- y ~ treatment*season +
f(spatial.field2, model = spde.pc) +
f(start,model = "rw2") +
f(individual,model = "iid") +
f(treatment_n,model = "rw2")
Mod <- inla(formula,
data = inla.stack.data(Stack, spde=spde.pc),
family = "lognormal",
control.compute = list(cpo = TRUE, config = TRUE),
control.predictor = list(A = inla.stack.A(Stack),
compute = T))
What I would like to get is the means with CrI's like shown below. To get an impression I calculated them out of group means of the dataset from fitted values (e.g. fit<-aggregate(mean ~ treatment + season, data = bar_dist, FUN= "mean" ).
Thanks!
Ramona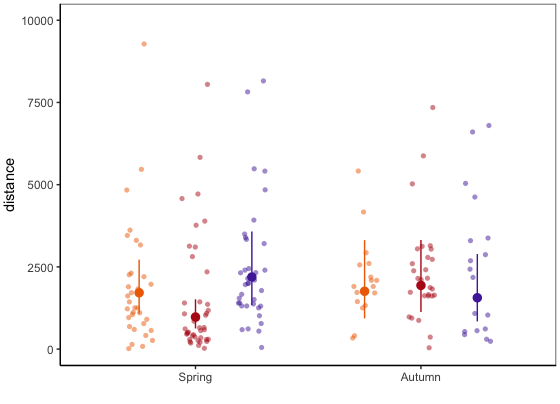

Reply all
Reply to author
Forward
0 new messages
