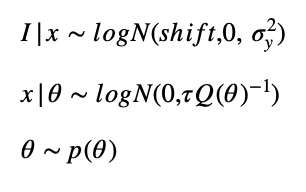shifted-lognormal distributed data
Amelie Liese
Finn Lindgren
--
You received this message because you are subscribed to the Google Groups "R-inla discussion group" group.
To unsubscribe from this group and stop receiving emails from it, send an email to r-inla-discussion...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/r-inla-discussion-group/21fb09f5-a709-47a6-9a38-a80e681940a6n%40googlegroups.com.
Amelie Liese
Hello,
thanks for your answer! The shift is known and doesn’t have to be estimated. I followed the link to the inlabru pages and have problems implementing my spde model with inlabru.
I have two main questions:
- Is it necessary to use inlabru to model a non-linear car-process or is it already implicit by the matern-correlation so using r-inla spde model would be sufficient?
- How does joining different simulated fields as iid realization work in inlabru?
Here are my model assumptions:
I want to fit a model to simulated cosmic matter-density-field data which is lognormal distributed. This data consists only of one data column („I“ density) and cartesian coordinates (on a unit sphere) columns:
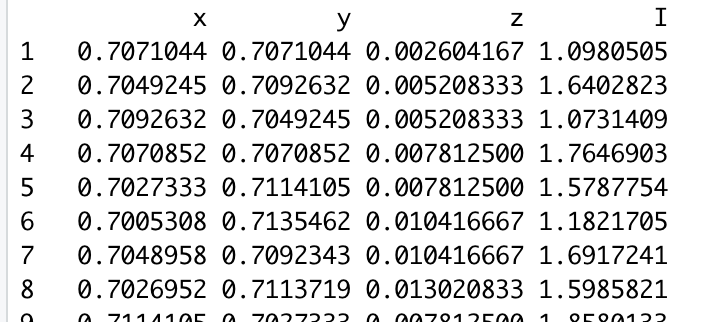
My model assumptions are:
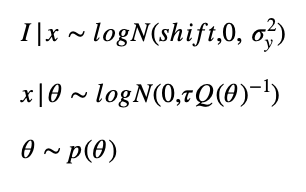
I assume, that the spatial process which generates the matter-density distribution can be described by a car-model which can be implemented by an spde-matern model for the parameters alpha = 2 and d = 1.
Therefore I assume that the linear predictor is the car-process over the neighborhood of x_s:
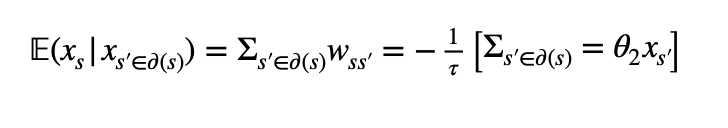
I have tried to model a non-linear car-process (gravitation process) over the neighborhood of x_s with inlabru and to define the latent field as lognormal distributed to better grasp the skewness of the data.
I also tried to join several simulated matter-density fields into one data sets, where the simulations should be modeled as iid realizations of the same process.
My code for inlabru:
(I only use window slices of the field not the whole sphere.)
## creating mesh with max.edge equals the size of prior correlation length until correlation 0.05
mesh <- inla.mesh.2d(loc = loc.cartesian, cutoff = 0.003, max.edge = c(0.005,0.01))
rep.id <- rep(1:n_maps, each = nrow(loc.cartesian))
x.loc <- rep(loc.cartesian[, 1], n_maps)
y.loc <- rep(loc.cartesian[, 2], n_maps)
z.loc <- rep(loc.cartesian[, 3], n_maps)
A <- inla.spde.make.A(mesh = mesh, loc = cbind(x.loc, y.loc,z.loc),
group = rep.id)
sigma0 <- 0.07. ## prior assumption of latent field standard deviation
range0 <- 0.005492295
lkappa0 <- log(8)/2 - log(range0)
ltau0 <- 0.5*log(1/(4*pi)) - log(sigma0) - lkappa0
spde <- inla.spde2.matern(mesh, alpha = 2, prior.variance.nominal = sigma0,
prioir.range.nomial = range0, prior.tau = exp(lkappa0),
prior.kappa = exp(lkappa0))
mesh.index <- inla.spde.make.index(name = "field",
n.spde = spde$n.spde,
n.group = n_maps)
obs <- as.vector(unlist(windows))
stack <- inla.stack( data = list(I = obs),
A = list(A,1),
effects = list(mesh.index, Intercept = rep(1, n_loc.cartesian*n_maps)),
tag = "est")
## cmp formula with spde(field, …) gives me an error message of not fitting lengths of objects,
## cmp formula without spde(field, …) can be fitted with bra but leads to non-plausible results.
cmp <- I ~ mySmooth(I, model = spde) + Intercept(1)
+ spde(field, model = spde, group = field.group, control.group = list(model = "iid"))
fit <- bru(components = cmp,
like(
formula = I ~ .,
family = "lognormal",
data = inla.stack.data(stack, spde = spde),
domain = list(x = mesh)
),
options = list(control.inla = list(int.strategy = "eb"))
)
Finn Lindgren
I have two main questions:
- Is it necessary to use inlabru to model a non-linear car-process or is it already implicit by the matern-correlation so using r-inla spde model would be sufficient?
- How does joining different simulated fields as iid realization work in inlabru?
My model assumptions are:
My code for inlabru:
(I only use window slices of the field not the whole sphere.)
## creating mesh with max.edge equals the size of prior correlation length until correlation 0.05
mesh <- inla.mesh.2d(loc = loc.cartesian, cutoff = 0.003, max.edge = c(0.005,0.01))
sigma0 <- 0.07. ## prior assumption of latent field standard deviation
range0 <- 0.005492295
spde <- inla.spde2.pcmatern(mesh, alpha = 2, prior.sigma=c(sigma0, 0.5), prior.range=c(range0,0.5))
# For iid replications, better to use replicate than group (group is used for dependent models):
cmp <- ~ -1 + Intercept(1) + field(cbind(x.loc, y.loc, z.loc), model=spde, replicate = real)
# The "domain" argument is only used for point process observations, so I removed it from your call:
fit <- bru(components = cmp,
like(
formula = obs ~ .,
family = "lognormal",
data = df
),
options = list(inla.mode = "experimental", control.inla = list(int.strategy = "eb"))
)
Amelie Liese
Thanks a lot for your answer and the corrections! I am still not sure if I understood it right and have some further questions:
the non-linearity aspect I am referring to:
One could for example model this via a polynomial regression :
E(matter_dens_s) = beta_0 + beta_1*matter_dens_s’ + beta_2*(matter_dens_s’)^2
The expectation value of the matter density at one site s depends on the matter density of the neighboring sites s'. Here I assume non-linearity in that way: A higher matter density of neighboring sites s’ has a bigger effect on the expectation value of the matter density at one site s than lower matter densities.
Thats why I thought I should include
I ~ mySmooth(I, model = spde)
In the formula. But If I understand your answer correctly, I have to link the expectation of the matter-density only to the latent field via the field(…, spde,…) object. Is the kind of non-linearity I am referring to covered by the field() component? Or do I have to include it in another way?
I also found some code examples for modeling spatial autocorrelation with a site() component (https://inbo.github.io/tutorials/tutorials/r_inla/spatial.pdf)
I ~ lon + lat + site(main = coordinates, model = spde)
Here the spatial autocorrelation is not modeled as a latent process. I am wondering why this is necessary. If I understand your correctly, this should be covered by the field() component?
I am writing a term paper on it the SPDE Approach and would like to read more about the relation between SPDE model representations and special CAR cases. It would be great if you could tell me some literature where it is explained more explicit.
Finn Lindgren
--
You received this message because you are subscribed to the Google Groups "R-inla discussion group" group.
To unsubscribe from this group and stop receiving emails from it, send an email to r-inla-discussion...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/r-inla-discussion-group/d74130ac-54f1-42dc-b027-5d49f079e523n%40googlegroups.com.
Amelie Liese
Hi Finn,
Thank you for the link to the paper and for the explanations. I now realized my mistake! The kind of non-linearity I actually mean is a non-linear effect of the distance between two observations! This non-linearity is referring to the curved space due to gravitation which effect is decreasing non-linear with the distance. I understand why my suggestions of
I ~ mySmooth(I, model = spde)
and
I ~ lon + lat + site(main = coordinates, model = spde)
Are wrong in that case!
The topic of my term paper is to present an approach for specifying a valid joint model of the cosmic matter density maps. That’s why I want to present the SPDE - Matérn approach that you have developed.
I have some questions relating two your answer:
If I want to include a non-linear distance effect I have to generate the distances to neighboring sites as covariates and include this in the formula as:
y_i = Intercept(1) + first_neigh_dis_i + sec_neigh_dis_i + field(s_i) + e_i
but then I can’t be sure that I specified a valid joint model of the map?
On the other side, if I want to ensure that I specify a valid joint model I start from the global perspective where the matérn field is projected on a finite dimensional Hilbert space which is spanned by local piecewise linear basis functions. Is the linearity in the basis functions related to the parameters? And if so, why can´t I model a non-linear effect over the degrees of neighbors (first oder neighbor, sec oder neighbor, ... )?
Kind regards,
Amelie Liese