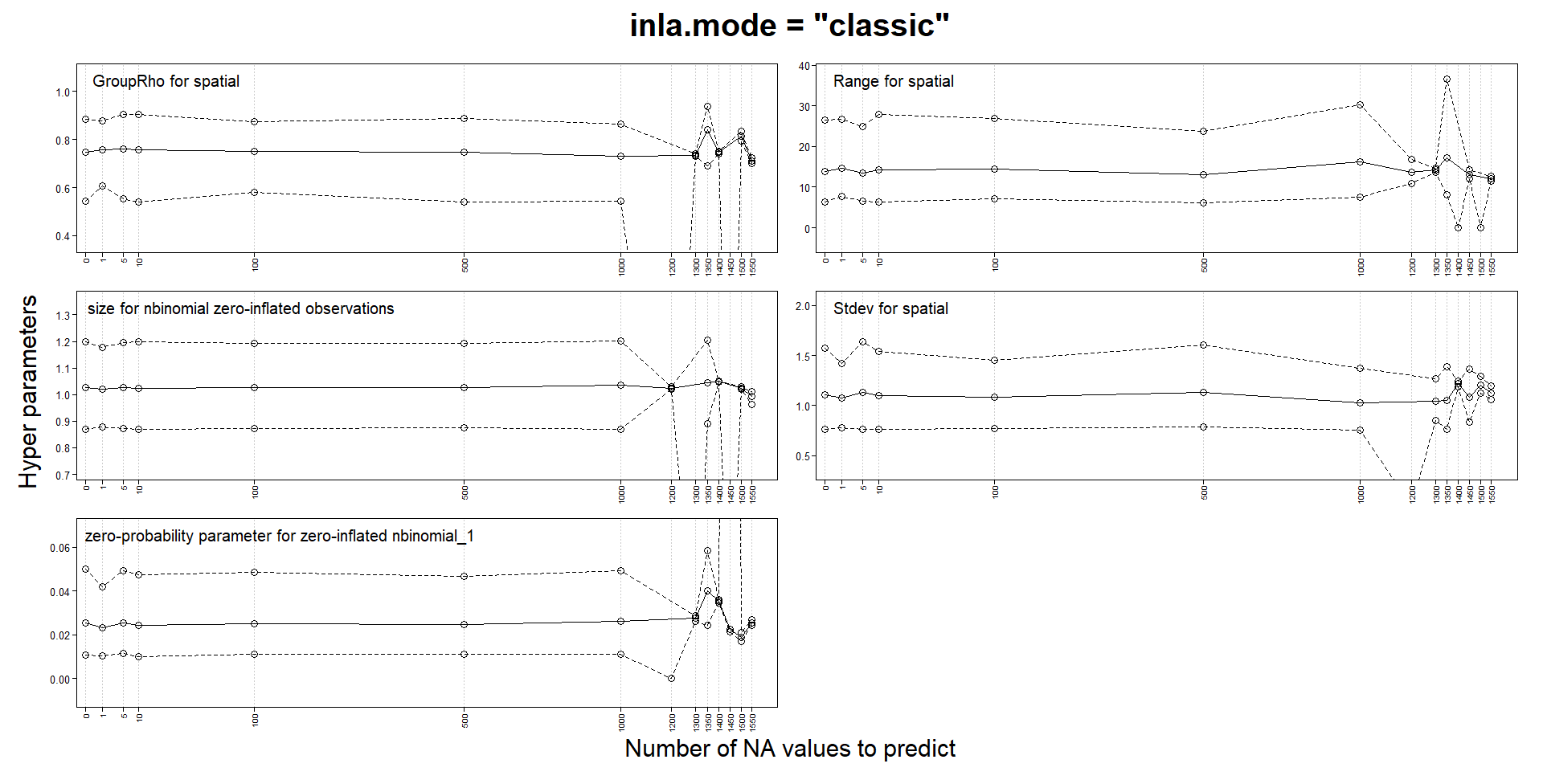
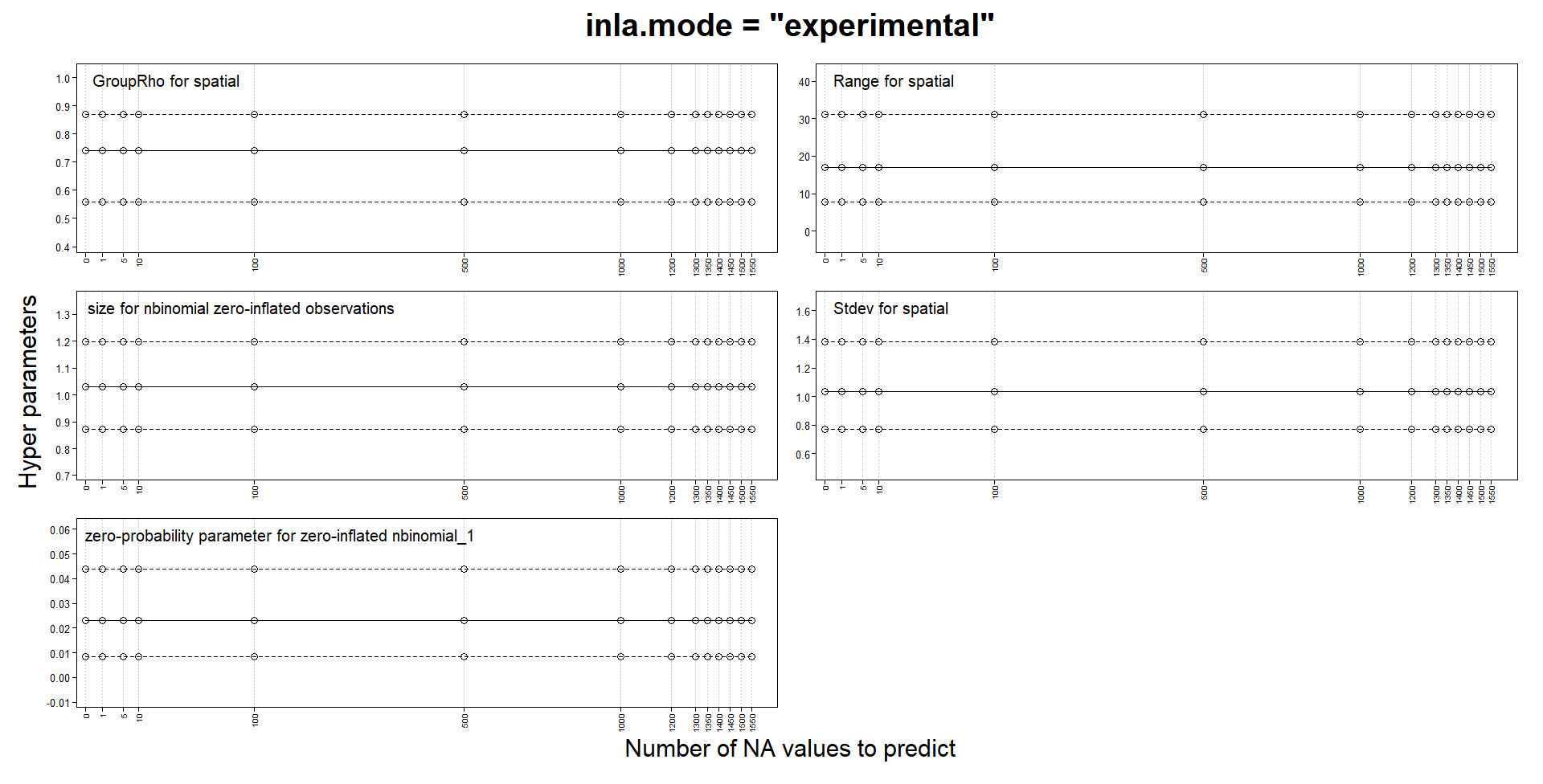
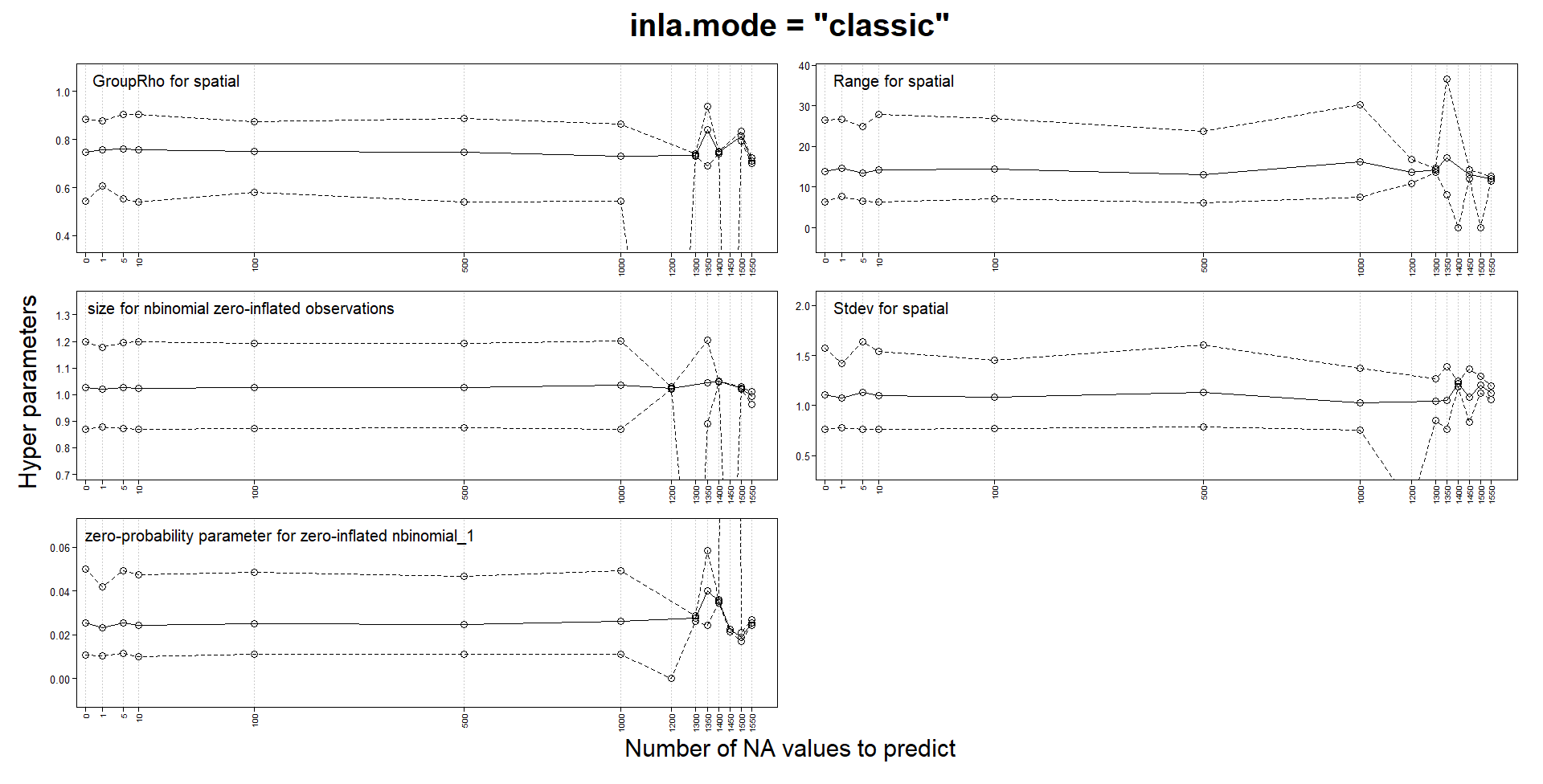
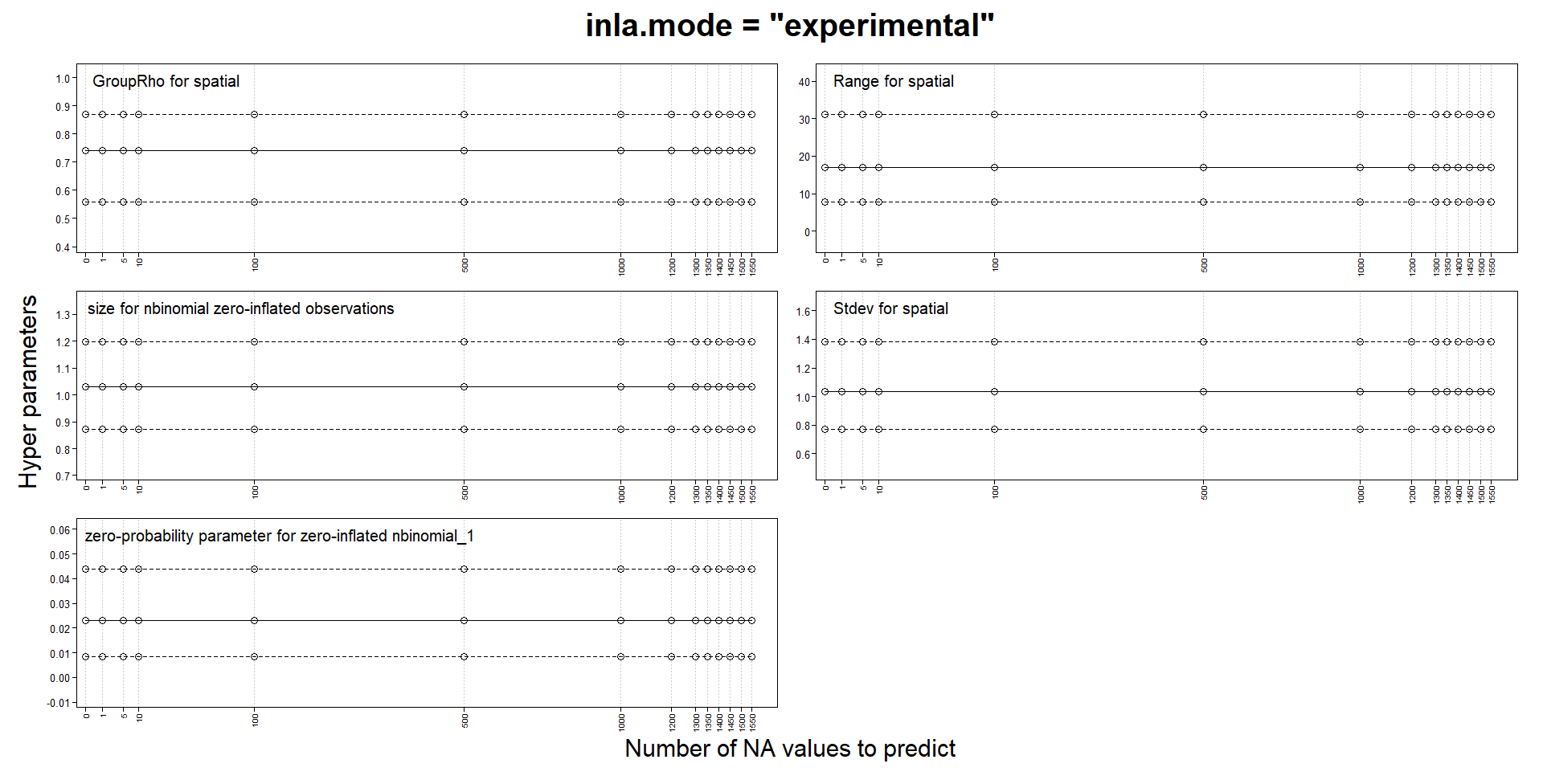
Classic mode changes estimates with predictions, but not experimental mode
140 views
Skip to first unread message
Francois R
Nov 25, 2021, 2:06:29 PM11/25/21
to R-inla discussion group
Hello INLA team
I am building spatiotemporal models with SPDE and an AR1 weekly component. My response is a zero-inflated count and I use a zero-inflated negative binomial model. My locations can change from week to week and the number of locations for certain weeks can be quite small.
Like several others, I noticed that parameters estimates could change when adding NA responses for prediction. In my case, they can change so much as to make the values NaN, Inf or sd of basically 0 or simply make INLA crash and run for a very long time. One of the advice for those issues is to try the "experimental" mode, so I compared the "classic" and the "experimental" mode on a tiny subset of my data. In this version, my model probably has to many fixed effects and I use a very coarse mesh to reduce computation times. I use a stack for estimation and another for prediction and I don't think I have made a mistake in their definition.
Thanks for your input and let me know if I should do further tests.
Francois Rousseu
Finn Lindgren
Nov 25, 2021, 2:23:30 PM11/25/21
to Francois R, R-inla discussion group
Hi Francois,
That’s a useful experiment! This matches my own observations when switching between classic and experimental mode for the automated inlabru package tests; in each case where the results changed when switching the mode, a closer inspection indicated that the experimental mode results were more accurate. The inlabru package tests now all use the experimental mode, but I haven’t decided on whether to add a default parameter setting for it yet, as I don’t want to deviate from he default inla settings unless needed. I believe Haavard and collaborators are still extending/testing the experimental mode a bit more, so will perhaps wait for that before switching the default. But I would currently recommend all spatial and spatio-temporal models (at least) to use it, regardless of whether problems with “classic” results are detected or not (as they are often only detected in extensive experiments such as yours, and apart from some temporary and obvious coding bugs that have been fixed, the experimental mode is much more stable), and if using classic mode, to do all predictions with posterior sampling instead. The experimental mode is also faster than classic mode.
Finn
On 25 Nov 2021, at 19:06, Francois R <francoi...@gmail.com> wrote:
--
You received this message because you are subscribed to the Google Groups "R-inla discussion group" group.
To unsubscribe from this group and stop receiving emails from it, send an email to r-inla-discussion...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/r-inla-discussion-group/f09d6c34-bac5-43cf-9ab1-340c9ee9b418n%40googlegroups.com.
Helpdesk
Nov 26, 2021, 2:51:04 AM11/26/21
to Finn Lindgren, Francois R, R-inla discussion group
Yes, very useful. It's a know feature, and it boil down to numerical
instabilities for almost singular matrices, which is controlled when
conditioning on data but gets worse when adding a (large) prediction
stack that is not just 'in-fill'. (pure) math says this is ok, but
reality is something else.
The 'experimental' mode is an internal rewrite (paper in progress) to
represent the linear predictor as deterministic of the 'model', which
thereby bypass these numerical issues and will provide also better
scalability and run-time (not uniformly, but in most cases).
this new internal representation comes with some restrictions, like we
cannot get the skewness correction for its marginal conditioned on each
hyperparameter but you get it when doing the integration. on the other
hand, the newly developed VB correction for the mean works just
great(!!!) within this framework and provide the road-map for how to
proceed with the developments in the future. there is/will-be-soon a new
report on arxiv
not all options are available for the 'experimental' mode, as during
rewrite only the 'useful' features was brought forward.
personally, I use the 'experimental' mode all the time, and the
numerical stability is better, run-time and memory is (much) better
almost uniformly, accuracy for the mean is better, etc... its still in
development, and will end up as the default option at some point. you
can enable it as default doing
library(INLA)
inla.setOption(inla.mode="experimental")
Best
Håvard
> > classic.pngexperimental.png
--
Håvard Rue
he...@r-inla.org
instabilities for almost singular matrices, which is controlled when
conditioning on data but gets worse when adding a (large) prediction
stack that is not just 'in-fill'. (pure) math says this is ok, but
reality is something else.
The 'experimental' mode is an internal rewrite (paper in progress) to
represent the linear predictor as deterministic of the 'model', which
thereby bypass these numerical issues and will provide also better
scalability and run-time (not uniformly, but in most cases).
this new internal representation comes with some restrictions, like we
cannot get the skewness correction for its marginal conditioned on each
hyperparameter but you get it when doing the integration. on the other
hand, the newly developed VB correction for the mean works just
great(!!!) within this framework and provide the road-map for how to
proceed with the developments in the future. there is/will-be-soon a new
report on arxiv
not all options are available for the 'experimental' mode, as during
rewrite only the 'useful' features was brought forward.
personally, I use the 'experimental' mode all the time, and the
numerical stability is better, run-time and memory is (much) better
almost uniformly, accuracy for the mean is better, etc... its still in
development, and will end up as the default option at some point. you
can enable it as default doing
library(INLA)
inla.setOption(inla.mode="experimental")
Best
Håvard
--
Håvard Rue
he...@r-inla.org
Helpdesk
Nov 26, 2021, 2:54:02 AM11/26/21
to Finn Lindgren, Francois R, R-inla discussion group
there is a discussion of this in the spde-book, or an section on how to
add predictions from a fitted model. I think section 7.4
Francois R
Nov 26, 2021, 7:52:24 AM11/26/21
to R-inla discussion group
Thanks to both of you for the input and advice. I will default to the experimental mode from now on and keep my eyes open for weird things.
François
Reply all
Reply to author
Forward
0 new messages