Can Qupath Import annotations?
1,836 views
Skip to first unread message
John Pan
Dec 10, 2018, 6:27:49 PM12/10/18
to QuPath users
Can Qupath Import annotations like NDPAs generated by NDPView2 or the XML file generated by Aperio Image Scope.
I attached some of the NDPA and XML files for reference.
I'm guessing this will require some basic scripting. Do I create an annotation and pass it a list of vertexes?
John
micros...@gmail.com
Dec 10, 2018, 8:40:55 PM12/10/18
to QuPath users
For Aperio I would check here first: https://github.com/qupath/qupath/issues/165s One of the links there is to a script in #61.
I don't know anything about the other file type.
John Pan
Jan 14, 2019, 5:24:22 PM1/14/19
to QuPath users
I modified that script to import NDPA files, but it was quite a bit more involved that I was expecting.

From this thread, I learned that NDPA files do not use a top-left centered coordinate system like Aperio does.

The Coordinate system that NDPAs use is relative to the physical slide center -- not even the scanned area center.
Additionally, it's not in pixels, it's in nanometers.
This value must be available somehow in QuPath thru open-slide when you open the slide, but I have no idea how to access it. @Pete?
In the absence of this value, I created a hack.
1. I manually drop a pin anywhere and name it "SlideCenter"
2. I manually open the NDPA file and find it in the XML. I manually set the X and Y coordinates to 1,1. (I used 0,0 before, but I experienced some unexplained bugs).
This is the physical center of the slide.

3. Reopen NDPVIEW and export annotation to CSV as opposed to NDPA. This is because the CSV option has ability to export to a Top-Left anchored coordinate system.
(I don't use this directly as the CSV export option only exports the annotation position, and not each point in an annotation, and the NDPA option has no option for Top-Left anchored coordinate system -- UGH).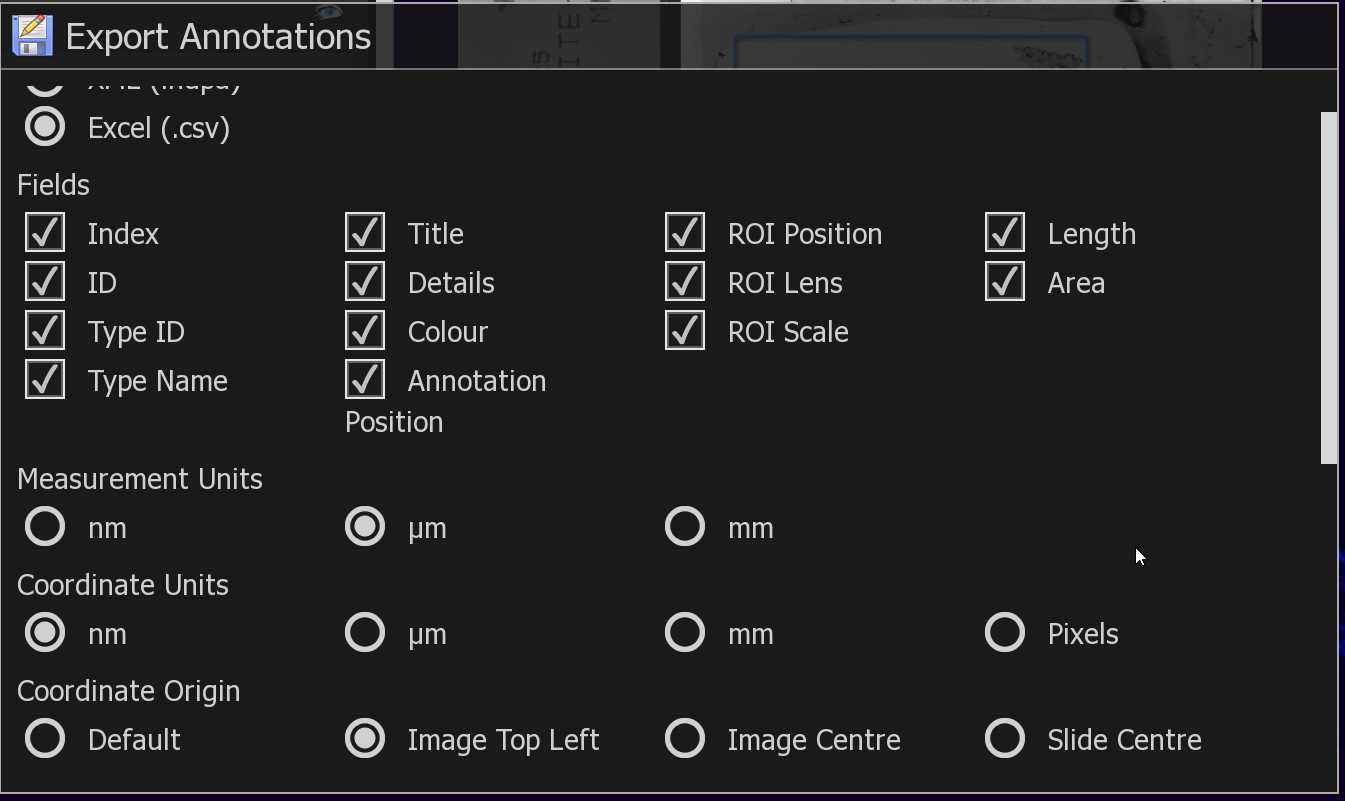

4. I now know the annotation point positions relative to slide center from the NDPA file, and the SlideCenter position from the TOP-LEFT from the .CSV file. -- all in Nanometers.
5. I run my script, which takes the NDPA, CSV file and pixels per micron information.
This is a terrible hack, but it works, and can be made much simpler if I could obtain the slide-center coordinates that Openslide must somehow use.
(excuse the poor code)
Message has been deleted
Pete
Jan 14, 2019, 5:39:46 PM1/14/19
to QuPath users
Wow, that does sound pretty complicated. I haven't encountered NDPA files before... perhaps I have been lucky.
Here's a terrible hack you can try:
def map = getCurrentImageData().getServer().osr.getProperties()
map.each { k, v ->
print k + ', ' + v
}It requests the current ImageServer, assumes (without checking) that it uses OpenSlide*, and accesses a private field... then gets the property map directly from OpenSlide. Hopefully it contains some info that helps.
*-Sometimes Bio-Formats can be used with .ndpi files also...
John Pan
Jan 14, 2019, 5:40:21 PM1/14/19
to QuPath users
This is a modification of DanaCase's script found here.
You will need to get the groovy-xml.jar as described here
John Pan
Jan 14, 2019, 5:41:34 PM1/14/19
to QuPath users
@Pete, can you provide similar code to get at bioformat's internals for SlideCenter?
Pete
Jan 14, 2019, 5:45:54 PM1/14/19
to QuPath users
Not as quickly... delving into Bio-Formats and OME-XML is a bigger job. It can be tackled independently of QuPath, i.e. create a new Bio-Formats ImageReader and set the ID to be the path to the image file, then... explore.
John Pan
Jan 16, 2019, 4:45:04 AM1/16/19
to QuPath users
I updated my code, it can now import directly from the NDPI without all the hacky Shenanigans due to being able to read coordinates from the openslide properties.
I left all the hacky stuff in as commented code in case anyone needs to reference it.
micros...@gmail.com
Jan 16, 2019, 9:34:11 AM1/16/19
to QuPath users
Thanks for the contribution!
John Pan
Jan 21, 2019, 8:20:55 PM1/21/19
to QuPath users
To anyone downloading these scripts.
The prints apparently massively slow them down. I'd recommend disabling them after understanding what they do.
The prints apparently massively slow them down. I'd recommend disabling them after understanding what they do.
Reply all
Reply to author
Forward
0 new messages
