Question about Digital Multiplexed IHC images
311 views
Skip to first unread message
Guray Akturk
Sep 11, 2018, 11:50:14 AM9/11/18
to QuPath users
Hi Everyone,
I need help with our digital multiplexing method.
We perform a multiplex IHC method in which we use same tissue section for staining with different antibodies. It's not a real multiplex immunostaining method. We use a chromogen (AEC) that we can easily bleach with ethanol and once we finish an immunostaining on a section, we scan the slide and save as a digital image. Afterwards we destain (bleach) the immunostaining with ethanol and continue with another antibody and do the same thing. After all we yield around 10 different antibody staining on the same section (individual digital images). We published this method in Science Immunology (DOI: 10.1126/sciimmunol.aaf6925).
Once we have digital images of multiple different immunostaining on the same section, we select a common region for all immunostainings to do registration and overlaying on Fiji (ImageJ). Once we export that region from 10 different digital slide, we save them as tiff in a folder and use Trakem2 plugin of Fiji to perfectly register them.
After the registration, we open registered images in Fiji and do image deconvolution, apply LUT and invert LUT, respectively to convert them to pseudoflourescent 8-bit images. After playing with the brightness setting, we use merge channels feature to overlay the images in Fiji.
After the overlaying, we yield an image including multiple channels for each antibody, but on a selected region from QuPath, not whole slide. We would like to open that overlaid image in QuPath and do cell segmentation but once we open that image file in QuPath, it is not possible for us to see different channels.
Other problem is whole slide registration and overlaying in Fiji: Resolution gets so high when we export the whole slide as tiff that, it becomes almost impossible to have enough memory. Is there any way to automatically export regions from QuPath to Fiji, do the registration and overlaying and stitch them together to use the memory effectively and create a whole slide image by stitching?
or, is there any way to import this multiple channel ROI onto the original image on QuPath?
Thanks
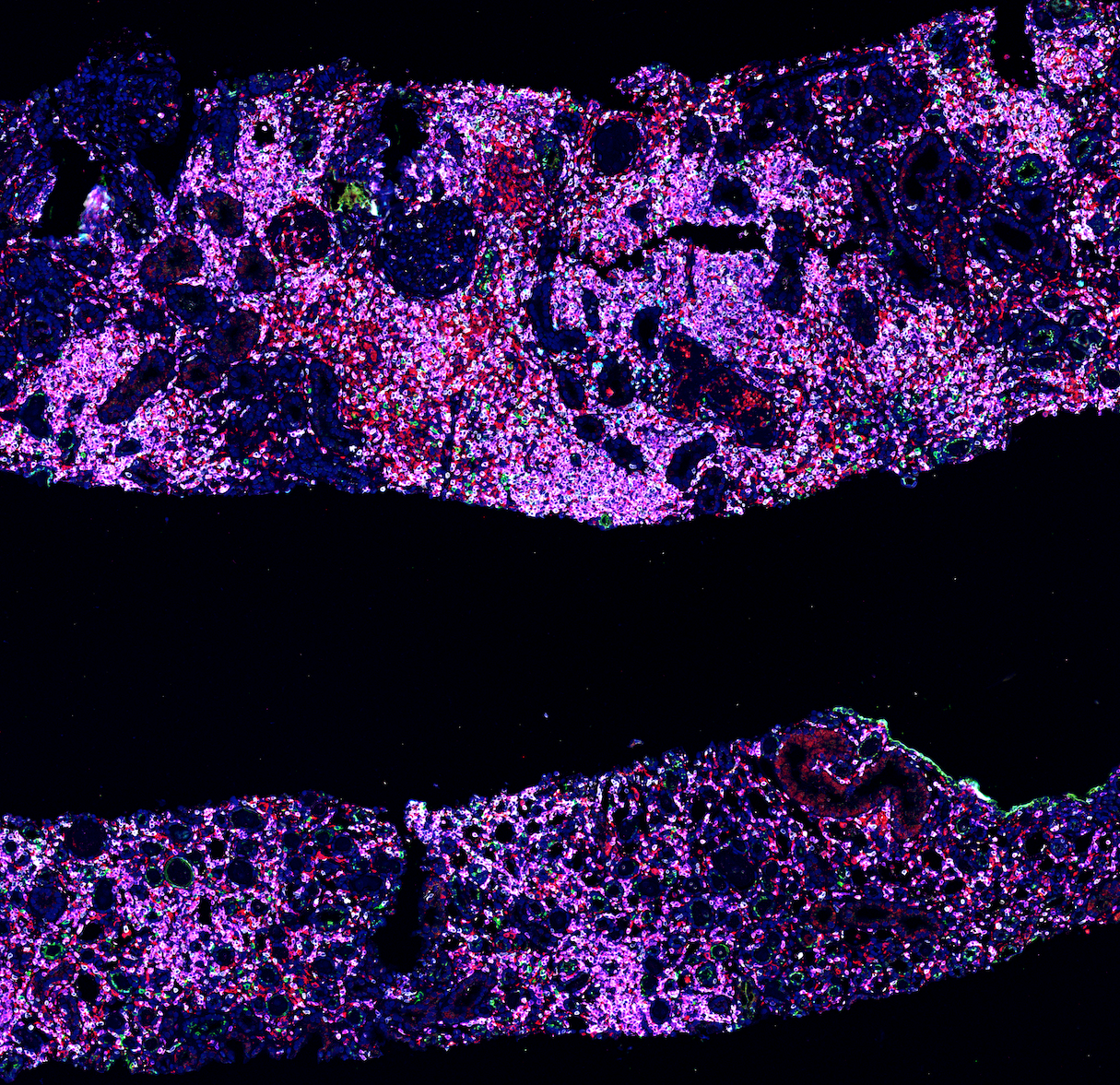
This is the result of digital registration and overlaying of different immunostains by using QuPath and Fiji together. Our ultimate plan is doing high-dimensional clustering by using QuPath detection measurements feature on multiple channels.
Pete
Sep 11, 2018, 12:15:52 PM9/11/18
to QuPath users
Hi Guray,
This is an interesting application; I've done bits and pieces of the work involved in performing dynamic stitching / stain separation / generation of pseudo-fluorescence images within QuPath, but lack the time, data, opportunity and immediate need to see it through to completion.
It should all be technically possible. Potentially you could do what you want through scripts/extensions/custom 'ImageServers', but it would be somewhat complex and interrelated with some other core changes I'm looking to make in QuPath over the next months (e.g. see https://petebankhead.github.io/qupath/tips/2018/08/06/multichannel-fluorescence.html ).
I think it would be great to integrate support for this in QuPath, and it is something I have been planning for a while (hence the 'bits and pieces'). Implementing this would take some work and is beyond what I can really describe here, but I'll send you a message in case it's something you want to discuss collaborating on in the future.
Regards,
Pete
Pete
Sep 11, 2018, 12:53:53 PM9/11/18
to QuPath users
Here's a link to the last related discussion: https://groups.google.com/d/msg/qupath-users/XNdaWK_9Ex4/VKHAbBGDBAAJ
Pete
Dec 27, 2018, 3:03:48 AM12/27/18
to QuPath users
Hi Guray,
I see that the QuPath + Fiji + TrakEM method is applied in the latest impressive preprint where you are coauthor: https://www.biorxiv.org/content/early/2018/12/20/503102.article-info
The description in the methods seems to contain an unfortunate error:
Images were processed by an experienced pathologist. Whole slide images obtained from scanned immunostains performed on the same tissue sections were opened simultaneously on our digital pathology image analysis software (QuPath 0.1.3).
I'm sure you can see why the description of QuPath by another group as 'our digital pathology image analysis software' is rather frustrating and potentially misleading, especially when combined with the absence of any citation. I hope that this will be corrected before the paper is published, and appropriate citations will be added for the software tools used developed by other researchers. All information is available, e.g.
Regards,
Pete
Guray Akturk
Apr 23, 2019, 11:53:00 AM4/23/19
to QuPath users
I was not aware of this problem and this post as well until microscopyra warned me today. I talked to the authors and they told me that they fixed the citation problem.
Please let me know if you notice something like this on any other papers in the future in which I am an author. I always do the citation but I would like to be more sure after this incident.
Sorry again, its on me that it got out of my attention.
Please let me know if you notice something like this on any other papers in the future in which I am an author. I always do the citation but I would like to be more sure after this incident.
Sorry again, its on me that it got out of my attention.
Reply all
Reply to author
Forward
0 new messages