Memory error with GLM
42 views
Skip to first unread message
john wisniewski
Oct 26, 2021, 11:43:32 AM10/26/21
to pystatsmodels
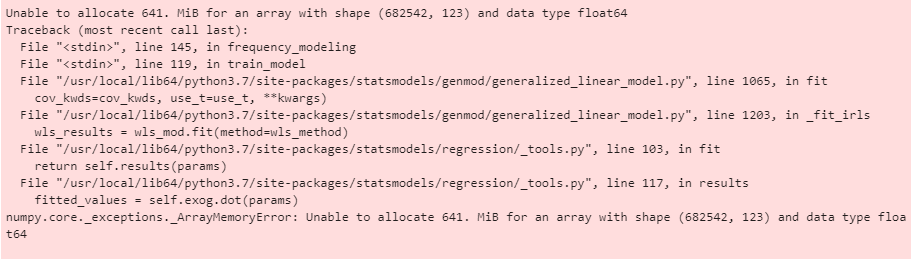
Hello, I've been working with sm.GLM (with family as Poisson) and keep getting memory errors when I try and fit my data. I have 122 features (using onehotencoder), and 682,542 rows. I've tried running it on my laptop (with 8gb ram), a desktop computer (with 16gb ram) and EMR within AWS.
I have been able to run the GLM with smaller subsections of the data (44 cols and 6000 rows). I can also run the full dataset with method = 'lbfgs' when I fit but this doesn't give the P-values and such.
I know this issue has been discussed before (I've read some of the other conversations) but I was wondering if this is actually the issue that's happening here or if there's something else going on. Am I actually having memory issues? Can provide more details if needed.
josef...@gmail.com
Oct 26, 2021, 1:10:43 PM10/26/21
to pystatsmodels
I guess you are just at the limit for in-memory computing on a regular computer.
The two main discussions were for out-of-memory computation and using sparse matrices for dummy variables.
In your case, it might be possible to save just enough memory to get it to run, but I'm not sure how much slack there is.
I find it a bit puzzling that numpy tries to allocate the full (682,542, 123) array for the dot product.
(maybe exog needs to be fortran contiguous for the linalg)
Did you check that you didn't use much memory already before creating the model?
Then we would need to check individual parts.
e.g. how much does memory consumption increase when creating the model?
If lbfgs works, then we would need to check whether or which hessian/cov_params computation will work.
IIRC fit(method= "xxx", max_start_irls=0, skip_hessian=True) should work for scipy optimizers
skip_hessian is mainly an internal keyword, and I'm not sure it works on the user sized.
It should prevent to automatically compute the hessian.
Also, You could try using discrete.Poisson instead of GLM-Poisson.
It's implemented specifically for the Poisson family and might need less memory. I never checked.
The big solution, out of memory and sparse matrices are likely not in the near future for statsmodels.
Josef
--
You received this message because you are subscribed to the Google Groups "pystatsmodels" group.
To unsubscribe from this group and stop receiving emails from it, send an email to pystatsmodel...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/pystatsmodels/c830ba0c-5fdc-4c36-82dc-d5f3d2a91762n%40googlegroups.com.
john wisniewski
Oct 27, 2021, 11:11:05 AM10/27/21
to pystatsmodels
Alright, thank you so much for your help! I'll look into measuring the memory at each stage and before I start running the code as well. Even if it still isn't possible to run, It's great to have some new directions to try, thank you.
Reply all
Reply to author
Forward
0 new messages
