Why FES calculated from meta-eabf is not smooth as that from WT-meta?
403 views
Skip to first unread message
Wei Zhang
Jun 8, 2021, 10:03:51 PM6/8/21
to PLUMED users
Dear all,
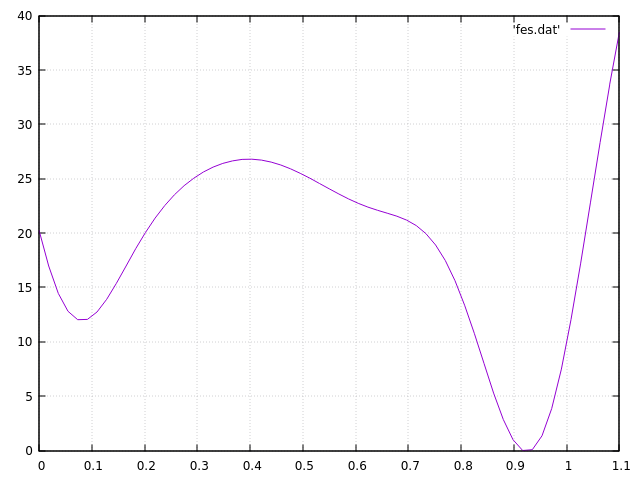
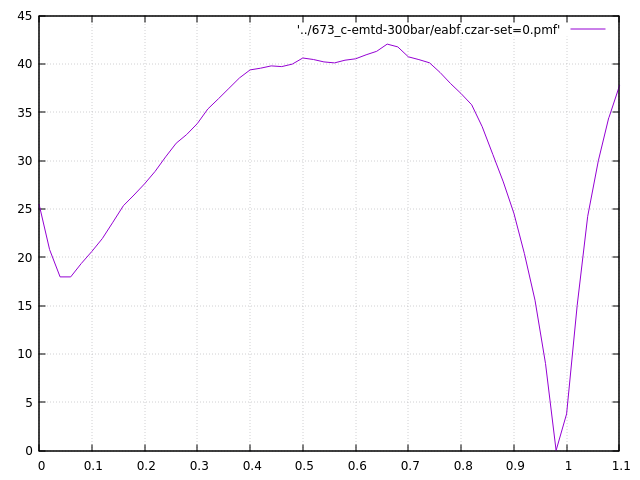
Recentlty, I was using Plumed-2v.6 with Lammps to calculate the PMF. The coordination number is CV, both WT-meta and meta-eabf method were used. However, I found the PMF calculated from calculated from meta-eabf is not smooth as that from WT-meta. Here follows the input scripts of these method and the output PMF figures.
1. Could anyone tell me how to change meta-eabf parameters to make the PMF line smooth?
2. How to estimate the error in free-energies with meta-eabf method?
#=========================meta-eabf=================
DRR ...
LABEL=eabf
ARG=ncoord
FULLSAMPLES=500
GRID_MAX=1.1
GRID_MIN=0.0
GRID_SPACING=0.02
FRICTION=8.0
TEXTOUTPUT
OUTPUTFREQ=5000
HISTORYFREQ=100000
TAU=0.5
TEMP=673
...
METAD ...
LABEL=mtd
ARG=eabf.ncoord_fict
SIGMA=0.1
GRID_MAX=3.0
GRID_MIN=-0.2
GRID_SPACING=0.02
HEIGHT=0.6
PACE=1000
TEMP=673
BIASFACTOR=8
...
uwalls: UPPER_WALLS ARG=eabf.ncoord_fict AT=1.10 KAPPA=1000000
lwalls: LOWER_WALLS ARG=eabf.ncoord_fict AT=0.00 KAPPA=1000000
#===================================meta========================
METAD ...
LABEL=mtd
ARG=ncoord
SIGMA=0.1
GRID_MAX=2.0
GRID_MIN=0
GRID_SPACING=0.02
# GRID_BIN=50
HEIGHT=0.6
PACE=1000
TEMP=673
BIASFACTOR=20
CALC_RCT
RCT_USTRIDE=10
...
#==================================================================


Haohao Fu
Jun 9, 2021, 4:53:24 AM6/9/21
to PLUMED users
Hi,
I noticed that you were using a similar Gaussian size for the MtD and WT-Meta-eABF simulations. I suspect that you want to use a smaller size for the WT-Meta-eABF simulation, as the biases from the ABF should dominate, and the MtD part only helps to guarantee a normal distribution of the sampling along the CV.
Good luck,
Haohao
Victor Hue
Jun 9, 2021, 5:38:40 AM6/9/21
to plumed...@googlegroups.com
Dear Zhang,
Hi Zhang. I'm learning how to use PLUMED in LAMMPS at the moment. I'm using the umbrella sampling method for my research.
Since you are using LAMMPS too, do you have any idea how can I use umbrella sampling in LAMMPS? From what I know, I have to run the simulations of each bias potential in parallel for different window stages, and from the trajectory produced, I need to combine all trajectories together, and run the simulation again with all the bias potentials.
I'm not sure how can I implemented this in LAMMPS. Specifically, how to run all simulation in parallel and how to combine the trajectories in LAMMPS.
Appreciate if you know anything about this. Thanks!
Regards,
Victor
--
You received this message because you are subscribed to the Google Groups "PLUMED users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to plumed-users...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/plumed-users/380ea5f9-4e04-4809-8464-f296325914a6n%40googlegroups.com.
Wei Zhang
Jun 9, 2021, 10:09:55 AM6/9/21
to PLUMED users
Dear Victor,
there are some examples of using lammps to do umbrella sampling with wham in the pdf file contained in the website(https://sites.udel.edu/it-rci/2018/02/28/applications-of-high-performance-computing-in-geochemistry/). Hope it will helps. (The pdf file is too larger to upload here.)
Good luck,
Wei
Victor Hue
Jun 10, 2021, 12:19:19 PM6/10/21
to plumed...@googlegroups.com
Dear Zhang,
Thanks for the website provided. I'm actually referring to the umbrella sampling using the PLUMED module (I'm currently using LAMMPS colvars module too) and try to compare both results. Thanks anyway!
Regards,
Victor
Thanks for the website provided. I'm actually referring to the umbrella sampling using the PLUMED module (I'm currently using LAMMPS colvars module too) and try to compare both results. Thanks anyway!
Regards,
Victor
To view this discussion on the web visit https://groups.google.com/d/msgid/plumed-users/c0483295-10b3-47ba-9a4e-a55dc90e54b9n%40googlegroups.com.
Chenghan Li
Sep 15, 2022, 2:21:26 PM9/15/22
to PLUMED users
Hi Wei,
I wonder if you used sum_hills to get the metad pmf. It uses a kernel density estimation by default which makes the pmf smooth. I am not sure about the abf thing.
Best,
Chenghan
Y Z
Apr 24, 2025, 6:25:50 AM4/24/25
to PLUMED users
Hello, every one. Sorry to disturb you guys. May I know how you compile the lammps with plumed. I compile with the following script
make mpi mode=shlib # build for whatever machine target you wish
make yes-all
make no-lib
make yes-MOLECULE
make yes-INTERLAYER
make yes-MANYBODY
make lib-plumed args="-b"
make yes-plumed
make mpi mode=shlib # build for whatever machine target you wish
make yes-all
make no-lib
make yes-MOLECULE
make yes-INTERLAYER
make yes-MANYBODY
make lib-plumed args="-b"
make yes-plumed
I can use plumed to run metadynamics successfully. But for the eabf, I always met the following error:
PLUMED: ERROR
PLUMED: I cannot understand line: DRR LABEL=eabf ARG=phi,psi FULLSAMPLES=500 GRID_MIN=-3.14,-3.14 GRID_MAX=3.14,3.14 GRID_BIN=180,180 FRICTION=8.0,8.0 TAU=0.5,0.5 OUTPUTFREQ=50000 HISTORYFREQ=500000
PLUMED: Maybe a missing space or a typo?
It seems that the plumed can not recognize the DRR command.
PLUMED: I cannot understand line: DRR LABEL=eabf ARG=phi,psi FULLSAMPLES=500 GRID_MIN=-3.14,-3.14 GRID_MAX=3.14,3.14 GRID_BIN=180,180 FRICTION=8.0,8.0 TAU=0.5,0.5 OUTPUTFREQ=50000 HISTORYFREQ=500000
PLUMED: Maybe a missing space or a typo?
It seems that the plumed can not recognize the DRR command.
Thanks a lot
Istvan Kolossvary
Apr 26, 2025, 2:11:19 PM4/26/25
to plumed...@googlegroups.com
Hi,
I am pretty sure that you installed the standard plumed, which does not include the DRR module. You have to build plumed from source (straightforward procedure, instructions on the website) and make sure that when you run ./configure you include --enable-boost_serialization and --enable-modules=drr on the command line.
Hope this helps,
Istvan
--
You received this message because you are subscribed to the Google Groups "PLUMED users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to plumed-users...@googlegroups.com.
To view this discussion visit https://groups.google.com/d/msgid/plumed-users/060c861d-2cce-45d1-8680-555bfcfea0f2n%40googlegroups.com.
Gourav Chakraborty
Jun 10, 2025, 7:44:28 AM6/10/25
to PLUMED users
Tell me one thing, how was the unbiasing done here? Is there any script that was used to get the resulting unbiased PMF? I am trying to implement a similar thing, with MetaD-eABF, but not sure how to get the PMF, coz both Meta and DRR give their own PMF profiles right? Which one to use?
Snow Summer
Jun 10, 2025, 11:28:54 AM6/10/25
to PLUMED users
Hi! You just need to integrate the .czar.grad file using the "abf_integrate" program in https://github.com/Colvars/colvars/tree/master/colvartools, as described in the plumed DRR manual (https://www.plumed.org/doc-v2.9/user-doc/html/_d_r_r.html).
Gourav Chakraborty
Jun 12, 2025, 2:20:16 AM6/12/25
to PLUMED users
Hello Everyone,
I tried one particular change in the WT-Meta-eABF code, please tell me if I am wrong here, instead of applyed WT-Meta on the fictitious particle, I used it on the actual CV. The change was significantly better, with eABF czar based PMF showing much higher phase space sampling. However, there is a downside to that, the obtained PMF values were substantially lower, mainly because CZAR was not able to account for the biasness introded by WT-Meta on the actual CV. However, my question is this, in the COLVAR script, I can get the ebaf.bias and meta.bias. Is it possible to use these to get the actual PMF?
I tried one particular change in the WT-Meta-eABF code, please tell me if I am wrong here, instead of applyed WT-Meta on the fictitious particle, I used it on the actual CV. The change was significantly better, with eABF czar based PMF showing much higher phase space sampling. However, there is a downside to that, the obtained PMF values were substantially lower, mainly because CZAR was not able to account for the biasness introded by WT-Meta on the actual CV. However, my question is this, in the COLVAR script, I can get the ebaf.bias and meta.bias. Is it possible to use these to get the actual PMF?
Reply all
Reply to author
Forward
0 new messages
