Applying rigid registration of two MR images using plastimatch warp
Alexandra B
Dear plastimatch community,
I am trying to apply a rigid registration (done with Velocity) of two MR images using the plastimatch warp command.
My original data is in dicom type (MRI and rigid registration object exported from Velocity). As a first step, I have used the command plastimatch convert, to convert the dicom files into mha.
The dicom
rigid registration has been performed using Velocity and exported in dcm
format. We are then trying to use this to rigidly align our images by
using the transformation matrix from the dicom rigid registration object
with the tag (3006,00C6) found in the dicom header.
My question is: Which convention for the Euler angles does plastimatch warp use?
I have tried
many conventions already, but my result has never been good enough. I.e.
the result differs from the rigidly aligned images directly exported
from velocity.
The best result is received using the following:
alpha = arctan(R_32/R_33); beta = arctan(-R_12/R_11); gamma = arctan(-R_23/sqrt(1-(R_23)^2)),
where the rotation matrix is simply given by [R_11 R_12 R_13; R_21 R_22 R_23; R_31 R_32 R_33].
The rotation parameters
were then the negative values of the Euler angles (-alpha, -beta,
-gamma) and the translation parameters were the negative translation vector entries from the transformation matrix (last
column of the 4x4 matrix).
I have chosen the fixed parameters to be (0 0 0).
Which changes do I have to make? Am I missing something straightforward?
I suspect it could have something to do with the image orientation of my MRI data. I have found that from the tag (0020,0037) in the dicom header of the MR image, I receive a rotation matrix as follows: R = [1 0 0; 0 0 -1; 0 1 0] (direction cosines).In the 4D Slicer VV, I receive a transformation matrix, which includes this rotation, but also a translation (see attached image). This translation is derived by computing the following:
origin – R * origin.
Could anyone tell me if this has an influence on the rigid registration using plastimatch warp? Do you have any experience in where this might be coming from?
Thank you for your help and best regards,
Alexandra
Attachments:
Here I have attached the rigid transformation matrix from the dicom tag (3006,00C6) for one of my cases:
.png?part=0.2&view=1)
Here the transformation matrix for the MRI data as seen in VV:
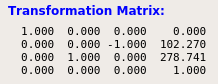
For the plastimatch warp command I then tried the following txt parameter file:

Sharp, Gregory C.
Hi Alexandra,
The Euler angles are defined by ITK and are not documented. I will take a look at the source code and get back to you.
The rotation matrix of VV is a conversion from IEC to DICOM coordinates. IEC is LSA whereas DICOM is LPS. The REG object should be DICOM: LPS and mm.
I'll note a few more things which may help. (Or may confuse.)
First, you can use plastimatch xf-convert to convert your ITK versor transform into a DICOM REG. This should help with debugging. Second, you can consider using the ITK Affine transform instead. Here is an example:
#Insight Transform File V1.0
#Transform 0
Transform: AffineTransform_double_3_3
Parameters: 1 0 0 0 1 0 0 0 1 20 40 60
FixedParameters: 0 0 0
I didn't fully understand your last question. Are you asking about registration or warping?
Greg
From: plast...@googlegroups.com <plast...@googlegroups.com>
On Behalf Of Alexandra B
Sent: Tuesday, November 22, 2022 11:17 AM
To: Plastimatch <plast...@googlegroups.com>
Subject: [Plastimatch] Applying rigid registration of two MR images using plastimatch warp
External Email - Use Caution

Here the transformation matrix for the MRI data as seen in VV:

For the plastimatch warp command I then tried the following txt parameter file:

--
You received this message because you are subscribed to the Google Groups "Plastimatch" group.
To unsubscribe from this group and stop receiving emails from it, send an email to
plastimatch...@googlegroups.com.
To view this discussion on the web visit
https://groups.google.com/d/msgid/plastimatch/45c6342c-4d98-46e6-8fe8-1e1e54ea0da6n%40googlegroups.com.
The information in this e-mail is intended only for the person to whom it is addressed. If you believe this e-mail was sent to you in error and the e-mail contains patient information, please contact the Mass General Brigham Compliance HelpLine at https://www.massgeneralbrigham.org/complianceline .
Please note that this e-mail is not secure (encrypted). If you do not wish to continue communication over unencrypted e-mail, please notify the sender of this message immediately. Continuing to send or respond to e-mail after receiving this message means you understand and accept this risk and wish to continue to communicate over unencrypted e-mail.
Sharp, Gregory C.
Hi again,
Here is the follow up. The ITK versor is not Euler angles. Instead, it is an angle-axis format. So, for R=(Rx,Ry,Rz), R / ||R|| is the unit vector about which the rotation takes place, and ||R|| is the magnitude of rotation.
-Greg
Alexandra B
Sharp, Gregory C.
Hi Alexandra,
Euler angles specify three successive rotations about the x, y, and/or z axes. Angle/axis specifies a single rotation about an arbitrary axis. The following article might be useful.
First you calculate the angle (theta, using the formula in the question), next you calculate the axis (v, using the formula in the answer), and finally you multiply angle with axis. For your matrix, I got: 0.128340157672645 0.950733780849466 0.282210203953310.
To generate the REG from the tfm, you need to specify the fixed and moving images such that the correct UIDs are used. If you have the original DICOM directories fixed/ and moving/, it is like this:
plastimatch xf-convert --input versor.tfm --output-dicom DICOM-SRO/ --fixed-rcs fixed/ --moving-rcs moving/
To view this discussion on the web visit https://groups.google.com/d/msgid/plastimatch/fc44516e-ad86-4ed4-b4c2-70b143c82e03n%40googlegroups.com.
