Fractions in peptide shaker
39 views
Skip to first unread message
valentine....@gmail.com
Nov 9, 2018, 3:04:25 AM11/9/18
to PeptideShaker
Hello,
Is peptide shaker have a maximum number of fraction that it could handle ?
Is the fractions in the "fraction"tab represent all the fraction analyzed or only the ones where proteins have been identified and validated ?
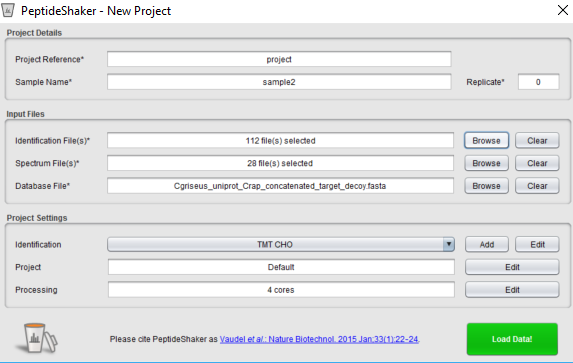
I have 28 fraction for one sample and only 16 are available in the fraction tab of peptide shaker despite a load of 28 spectrum files from the SearchGUI output file.
Thank you for the clarification,
Best regards,
Valentine
Harald Barsnes
Nov 9, 2018, 7:14:23 AM11/9/18
to PeptideShaker
Hi Valentine,
As far as I can remember the fraction tab should show all fractions, but for some of the tabs at the bottom only the validated results are displayed. Could you share a screenshot showing where you only have 16 out of 28 fractions?
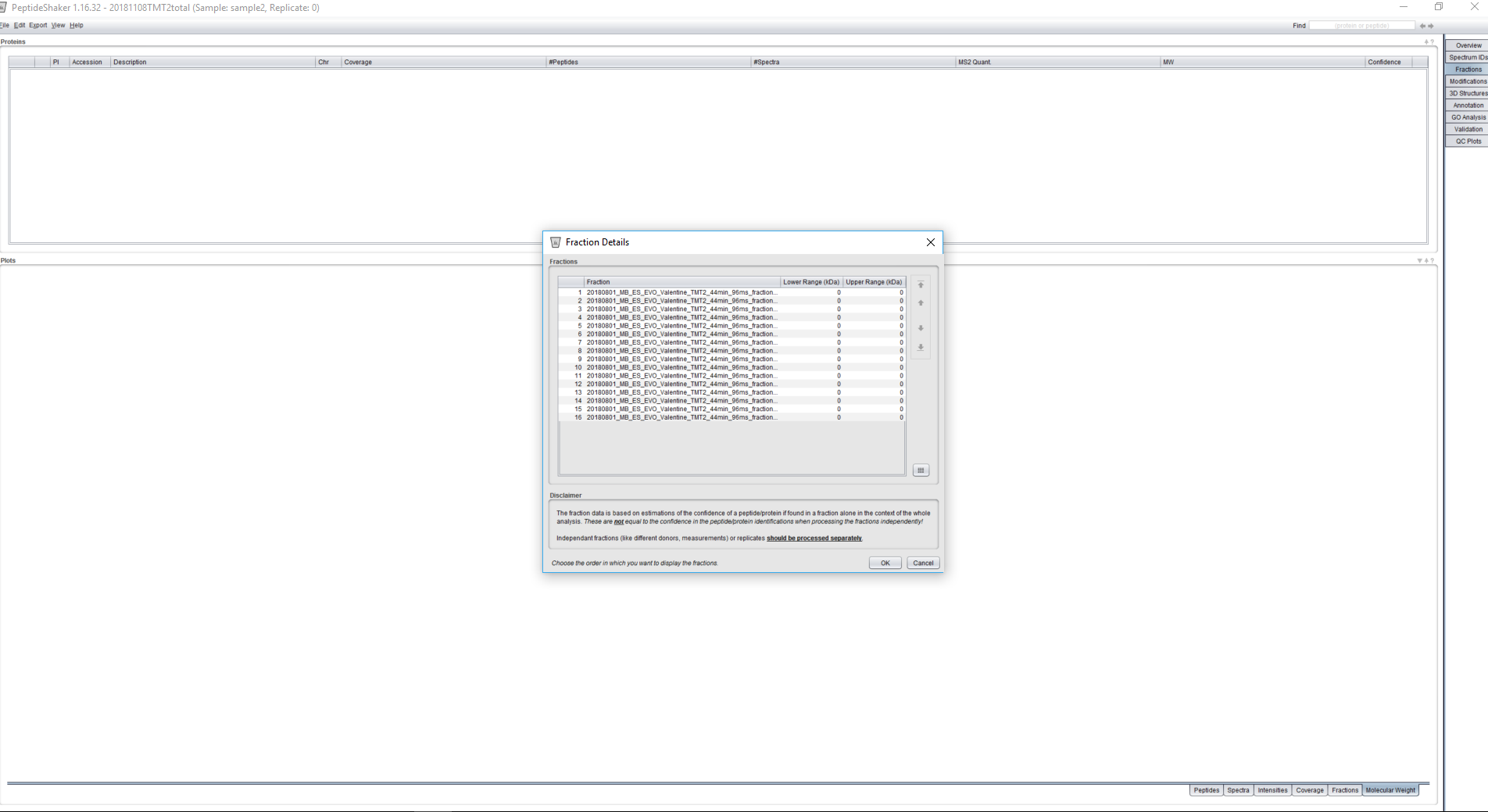
Perhaps you can also check the Fraction Details dialog available via Edit > Fraction Details to verify whether all the fractions are showing there?
Best regards,
Harald
valentine chevallier
Nov 9, 2018, 9:35:11 AM11/9/18
to peptide...@googlegroups.com
Hi Harald,
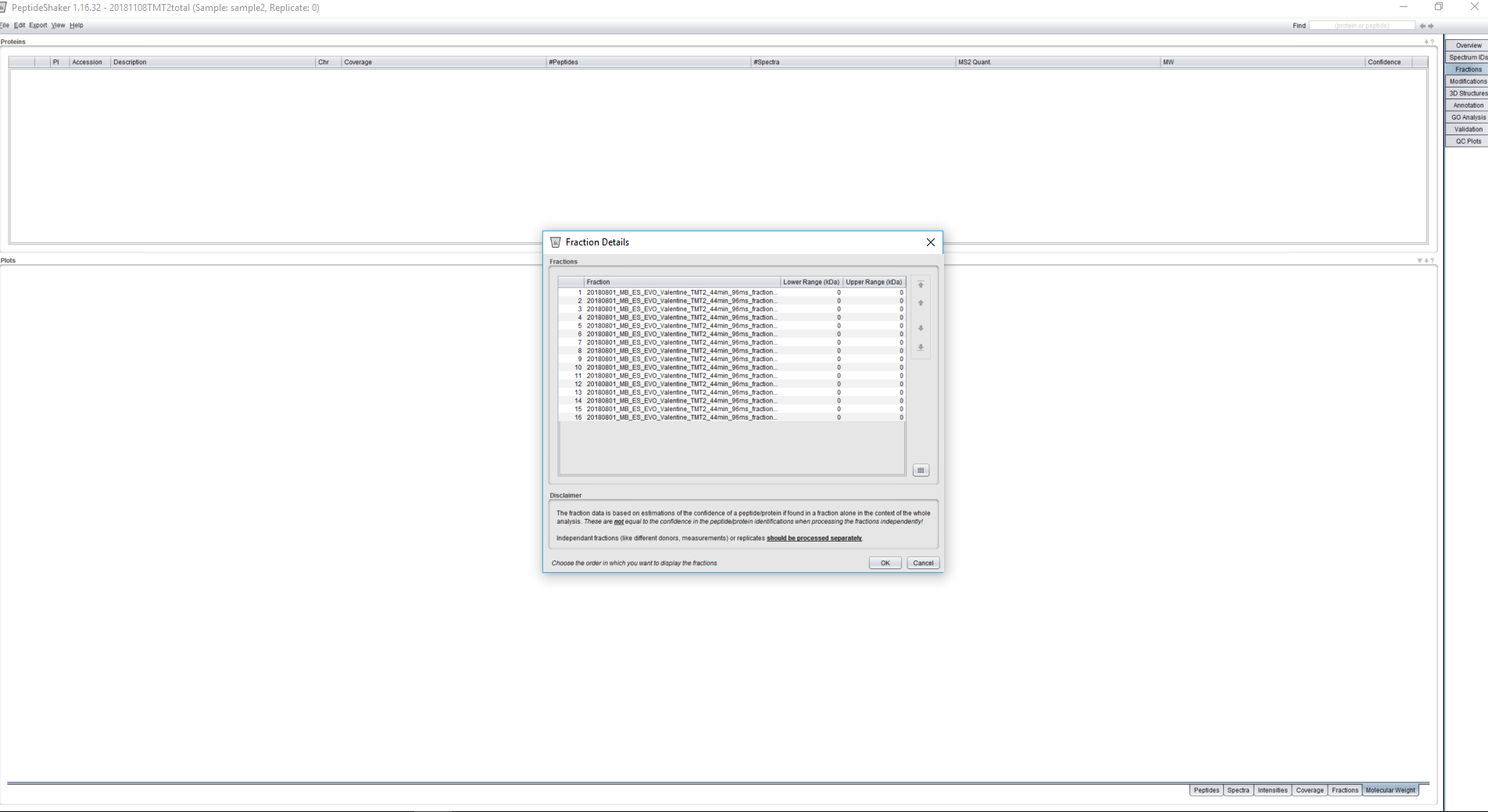
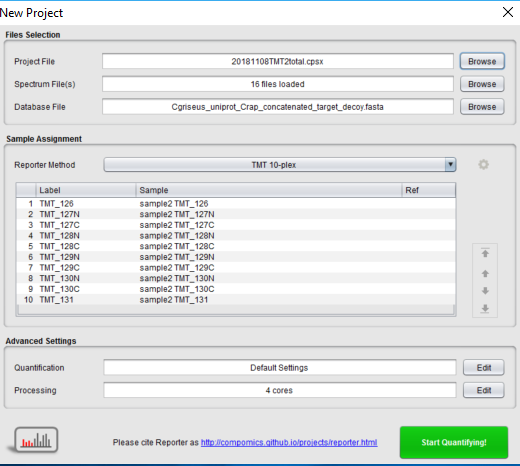
Before peptide shaker I have 28 spectrum files in my searchgui output :

then after the peptide shaker processing I have 16 fractions in the fraction tab and in the fraction details :

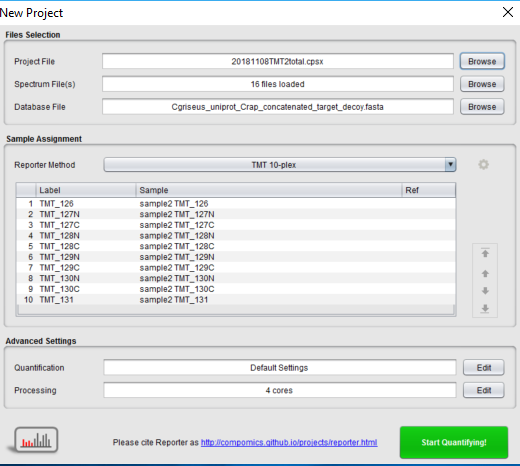
And then when I am uploading the project in reporter I have only 16 spectrum files :

I hope it helps,
Best regards,
Valentine
--
You received this message because you are subscribed to the Google Groups "PeptideShaker" group.
To unsubscribe from this group and stop receiving emails from it, send an email to peptide-shake...@googlegroups.com.
For more options, visit https://groups.google.com/d/optout.
Harald Barsnes
Nov 13, 2018, 9:48:22 AM11/13/18
to PeptideShaker
Hi Valentine,
Do you see any pattern with regards to which spectrum files that show up in the Fractions tab? And are you sure that there are identification results for all of the spectrum files?
Perhaps you could try loading only the search results of one of the spectrum files not showing up in the Fractions tab?
And I assume you have checked the SearchGUI or PeptideShaker logs to see if they contain any useful information?
Best regards,
Harald
On Friday, November 9, 2018 at 3:35:11 PM UTC+1, valentine chevallier wrote:
Hi Harald,Before peptide shaker I have 28 spectrum files in my searchgui output :then after the peptide shaker processing I have 16 fractions in the fraction tab and in the fraction details :And then when I am uploading the project in reporter I have only 16 spectrum files :I hope it helps,Best regards,Valentine
Le ven. 9 nov. 2018 à 13:14, Harald Barsnes a écrit :
--Hi Valentine,As far as I can remember the fraction tab should show all fractions, but for some of the tabs at the bottom only the validated results are displayed. Could you share a screenshot showing where you only have 16 out of 28 fractions?Perhaps you can also check the Fraction Details dialog available via Edit > Fraction Details to verify whether all the fractions are showing there?Best regards,Harald
On Friday, November 9, 2018 at 9:04:25 AM UTC+1, valentine.chevallier wrote:Hello,Is peptide shaker have a maximum number of fraction that it could handle ?Is the fractions in the "fraction"tab represent all the fraction analyzed or only the ones where proteins have been identified and validated ?I have 28 fraction for one sample and only 16 are available in the fraction tab of peptide shaker despite a load of 28 spectrum files from the SearchGUI output file.Thank you for the clarification,Best regards,Valentine
You received this message because you are subscribed to the Google Groups "PeptideShaker" group.
To unsubscribe from this group and stop receiving emails from it, send an email to peptide-shaker+unsubscribe@googlegroups.com.
valentine chevallier
Nov 13, 2018, 9:59:52 AM11/13/18
to peptide...@googlegroups.com
Hi harald,
I have found the issue in the log of peptide shaker : "More than 75% of the PSMs did not pass the import filters.Please verify that your peptide selection criteria are not too restrictive. Please verify that your precursor selection criteria are not too restrictive." Sorry I haven't checked before. So all the fraction didn't appears because of the filtering.
I imagine that default parameters didn't fit with a 11 plex TMT labelling run on QExactive?
Best regards,
Valentine
To unsubscribe from this group and stop receiving emails from it, send an email to peptide-shake...@googlegroups.com.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to the Google Groups "PeptideShaker" group.
To unsubscribe from this group and stop receiving emails from it, send an email to peptide-shake...@googlegroups.com.
Harald Barsnes
Nov 13, 2018, 10:24:44 AM11/13/18
to PeptideShaker
Hi Valentine,
Thanks the update. What does it say with regards to why the PSMs are excluded? You should have a percentage in each category?
> I imagine that default parameters didn't fit with a 11 plex TMT labelling run on QExactive?
Correct. You have to optimize the search parameters to fit the settings of the given instruments.
I would also recommend checking the import filters used. You can find these by clicking Edit after the identification settings in the PeptideShaker New Project dialog, then clicking "Show Advanced Settings" and finally click the Import Filters option.
Best regards,
Harald
valentine chevallier
Nov 13, 2018, 12:37:46 PM11/13/18
to peptide...@googlegroups.com
Dear Harald,
you can find below or in attachment the details. I was wondering that is I add TMT 11plex as a fixed modification it will takes in account the mass shift due to the label. But I should also change the range of isotope deviation in the method ?
you can find below or in attachment the details. I was wondering that is I add TMT 11plex as a fixed modification it will takes in account the mass shift due to the label. But I should also change the range of isotope deviation in the method ?
Thank you for your help,
Kind regards,
Valentine
777 identified spectra (78.5%) did not present a valid peptide. Sun Nov 11 08:29:59 CET 2018 994 of the best scoring peptides were excluded by the import filters: Sun Nov 11 08:29:59 CET 2018 - 21.8% peptide length less than 8 or greater than 30. Sun Nov 11 08:29:59 CET 2018 - 77.7% peptide presenting high mass or isotopic deviation. Sun Nov 11 08:29:59 CET 2018 Warning: More than 75% of the PSMs did not pass the import filters. Please verify that your peptide selection criteria are not too restrictive. Please verify that your precursor selection criteria are not too restrictive.
--
Harald Barsnes
Nov 14, 2018, 11:22:44 AM11/14/18
to PeptideShaker
Hi Valentine,
I don't see anything obviously wrong with your search parameters. And unless your instrument is very frequently picking the wrong isotope, the isotope deviation setting should not be vital.
I'd recommend loading smaller subsets of your search results to try to locate the problem. For example just the search results from a single search engine. Or even just the search results from a single mgf file. Starting with just a single search result file.
After loading the data go to the QC Plots tab (found in the upper right corner) and see if the provided plots can bring you closer to figuring out what is going on.
Best regards,
Harald
Reply all
Reply to author
Forward
0 new messages