FreeMarker template error in study.model...
Iago Giné Vázquez
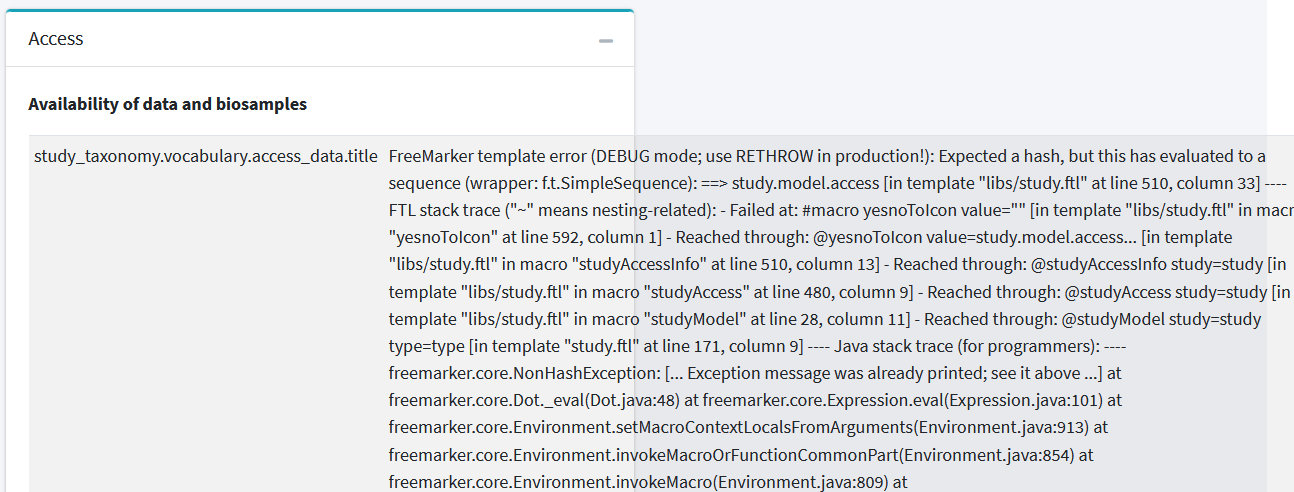
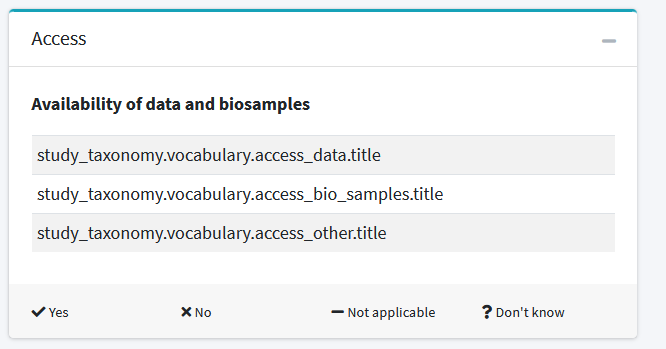
The full error of the first image:
```
study_taxonomy.vocabulary.access_data.title
FreeMarker template error (DEBUG mode; use RETHROW in production!): Expected a hash, but this has evaluated to a sequence (wrapper: f.t.SimpleSequence): ==> study.model.access [in template "libs/study.ftl" at line 510, column 33] ---- FTL stack trace ("~" means nesting-related): - Failed at: #macro yesnoToIcon value="" [in template "libs/study.ftl" in macro "yesnoToIcon" at line 592, column 1] - Reached through: @yesnoToIcon value=study.model.access... [in template "libs/study.ftl" in macro "studyAccessInfo" at line 510, column 13] - Reached through: @studyAccessInfo study=study [in template "libs/study.ftl" in macro "studyAccess" at line 480, column 9] - Reached through: @studyAccess study=study [in template "libs/study.ftl" in macro "studyModel" at line 28, column 11] - Reached through: @studyModel study=study type=type [in template "study.ftl" at line 171, column 9] ---- Java stack trace (for programmers): ---- freemarker.core.NonHashException: [... Exception message was already printed; see it above ...] at freemarker.core.Dot._eval(Dot.java:48) at freemarker.core.Expression.eval(Expression.java:101) at freemarker.core.Environment.setMacroContextLocalsFromArguments(Environment.java:913) at freemarker.core.Environment.invokeMacroOrFunctionCommonPart(Environment.java:854) at freemarker.core.Environment.invokeMacro(Environment.java:809) at freemarker.core.UnifiedCall.accept(UnifiedCall.java:83) at freemarker.core.Environment.visit(Environment.java:331) at freemarker.core.Environment.visit(Environment.java:373) at freemarker.core.Environment.invokeMacroOrFunctionCommonPart(Environment.java:872) at freemarker.core.Environment.invokeMacro(Environment.java:809) at freemarker.core.UnifiedCall.accept(UnifiedCall.java:83) at freemarker.core.Environment.visit(Environment.java:331) at freemarker.core.Environment.visit(Environment.java:373) at freemarker.core.Environment.invokeMacroOrFunctionCommonPart(Environment.java:872) at freemarker.core.Environment.invokeMacro(Environment.java:809) at freemarker.core.UnifiedCall.accept(UnifiedCall.java:83) at freemarker.core.Environment.visit(Environment.java:331) at freemarker.core.Environment.visit(Environment.java:337) at freemarker.core.Environment.visit(Environment.java:373) at freemarker.core.Environment.invokeMacroOrFunctionCommonPart(Environment.java:872) at freemarker.core.Environment.invokeMacro(Environment.java:809) at freemarker.core.UnifiedCall.accept(UnifiedCall.java:83) at freemarker.core.Environment.visit(Environment.java:331) at freemarker.core.Environment.visit(Environment.java:337) at freemarker.core.Environment.process(Environment.java:310) at freemarker.template.Template.process(Template.java:383) at org.springframework.web.servlet.view.freemarker.FreeMarkerView.processTemplate(FreeMarkerView.java:368) at org.springframework.web.servlet.view.freemarker.FreeMarkerView.doRender(FreeMarkerView.java:285) at org.springframework.web.servlet.view.freemarker.FreeMarkerView.renderMergedTemplateModel(FreeMarkerView.java:235) at org.springframework.web.servlet.view.AbstractTemplateView.renderMergedOutputModel(AbstractTemplateView.java:167) at org.springframework.web.servlet.view.AbstractView.render(AbstractView.java:304) at org.springframework.web.servlet.DispatcherServlet.render(DispatcherServlet.java:1286) at org.springframework.web.servlet.DispatcherServlet.processDispatchResult(DispatcherServlet.java:1041) at org.springframework.web.servlet.DispatcherServlet.doDispatch(DispatcherServlet.java:984) at org.springframework.web.servlet.DispatcherServlet.doService(DispatcherServlet.java:901) at org.springframework.web.servlet.FrameworkServlet.processRequest(FrameworkServlet.java:970) at org.springframework.web.servlet.FrameworkServlet.doGet(FrameworkServlet.java:861) at javax.servlet.http.HttpServlet.service(HttpServlet.java:687) at org.springframework.web.servlet.FrameworkServlet.service(FrameworkServlet.java:846) at javax.servlet.http.HttpServlet.service(HttpServlet.java:790) at org.eclipse.jetty.servlet.ServletHolder.handle(ServletHolder.java:755) at org.eclipse.jetty.servlet.ServletHandler$CachedChain.doFilter(ServletHandler.java:1617) at org.obiba.mica.web.filter.ClickjackingHttpHeadersFilter.doFilter(ClickjackingHttpHeadersFilter.java:41) at org.eclipse.jetty.servlet.ServletHandler$CachedChain.doFilter(ServletHandler.java:1604) at com.codahale.metrics.servlet.AbstractInstrumentedFilter.doFilter(AbstractInstrumentedFilter.java:97) at org.eclipse.jetty.servlet.ServletHandler$CachedChain.doFilter(ServletHandler.java:1604) at org.obiba.mica.config.WebConfiguration$NoTraceFilter.doFilter(WebConfiguration.java:298) at org.eclipse.jetty.servlet.ServletHandler$CachedChain.doFilter(ServletHandler.java:1604) at org.eclipse.jetty.websocket.server.WebSocketUpgradeFilter.doFilter(WebSocketUpgradeFilter.java:226) at org.eclipse.jetty.servlet.ServletHandler$CachedChain.doFilter(ServletHandler.java:1604) at org.obiba.shiro.web.filter.AuthenticationFilter.doFilterInternal(AuthenticationFilter.java:158) at org.springframework.web.filter.OncePerRequestFilter.doFilter(OncePerRequestFilter.java:107) at org.eclipse.jetty.servlet.ServletHandler$CachedChain.doFilter(ServletHandler.java:1604) at org.springframework.web.filter.RequestContextFilter.doFilterInternal(RequestContextFilter.java:99) at org.springframework.web.filter.OncePerRequestFilter.doFilter(OncePerRequestFilter.java:107) at org.eclipse.jetty.servlet.ServletHandler$CachedChain.doFilter(ServletHandler.java:1604) at org.springframework.web.filter.HttpPutFormContentFilter.doFilterInternal(HttpPutFormContentFilter.java:109) at org.springframework.web.filter.OncePerRequestFilter.doFilter(OncePerRequestFilter.java:107) at org.eclipse.jetty.servlet.ServletHandler$CachedChain.doFilter(ServletHandler.java:1604) at org.springframework.web.filter.HiddenHttpMethodFilter.doFilterInternal(HiddenHttpMethodFilter.java:93) at org.springframework.web.filter.OncePerRequestFilter.doFilter(OncePerRequestFilter.java:107) at org.eclipse.jetty.servlet.ServletHandler$CachedChain.doFilter(ServletHandler.java:1604) at org.springframework.web.filter.CharacterEncodingFilter.doFilterInternal(CharacterEncodingFilter.java:197) at org.springframework.web.filter.OncePerRequestFilter.doFilter(OncePerRequestFilter.java:107) at org.eclipse.jetty.servlet.ServletHandler$CachedChain.doFilter(ServletHandler.java:1604) at org.eclipse.jetty.servlet.ServletHandler.doHandle(ServletHandler.java:545) at org.eclipse.jetty.server.handler.ScopedHandler.handle(ScopedHandler.java:143) at org.eclipse.jetty.security.SecurityHandler.handle(SecurityHandler.java:590) at org.eclipse.jetty.server.handler.HandlerWrapper.handle(HandlerWrapper.java:127) at org.eclipse.jetty.server.handler.ScopedHandler.nextHandle(ScopedHandler.java:235) at org.eclipse.jetty.server.session.SessionHandler.doHandle(SessionHandler.java:1607) at org.eclipse.jetty.server.handler.ScopedHandler.nextHandle(ScopedHandler.java:233) at org.eclipse.jetty.server.handler.ContextHandler.doHandle(ContextHandler.java:1297) at org.eclipse.jetty.server.handler.ScopedHandler.nextScope(ScopedHandler.java:188) at org.eclipse.jetty.servlet.ServletHandler.doScope(ServletHandler.java:485) at org.eclipse.jetty.server.session.SessionHandler.doScope(SessionHandler.java:1577) at org.eclipse.jetty.server.handler.ScopedHandler.nextScope(ScopedHandler.java:186) at org.eclipse.jetty.server.handler.ContextHandler.doScope(ContextHandler.java:1212) at org.eclipse.jetty.server.handler.ScopedHandler.handle(ScopedHandler.java:141) at org.eclipse.jetty.server.handler.gzip.GzipHandler.handle(GzipHandler.java:767) at org.eclipse.jetty.server.handler.HandlerWrapper.handle(HandlerWrapper.java:127) at org.eclipse.jetty.server.Server.handle(Server.java:500) at org.eclipse.jetty.server.HttpChannel.lambda$handle$1(HttpChannel.java:383) at org.eclipse.jetty.server.HttpChannel.dispatch(HttpChannel.java:547) at org.eclipse.jetty.server.HttpChannel.handle(HttpChannel.java:375) at org.eclipse.jetty.server.HttpConnection.onFillable(HttpConnection.java:270) at org.eclipse.jetty.io.AbstractConnection$ReadCallback.succeeded(AbstractConnection.java:311) at org.eclipse.jetty.io.FillInterest.fillable(FillInterest.java:103) at org.eclipse.jetty.io.ChannelEndPoint$2.run(ChannelEndPoint.java:117) at org.eclipse.jetty.util.thread.strategy.EatWhatYouKill.runTask(EatWhatYouKill.java:336) at org.eclipse.jetty.util.thread.strategy.EatWhatYouKill.doProduce(EatWhatYouKill.java:313) at org.eclipse.jetty.util.thread.strategy.EatWhatYouKill.tryProduce(EatWhatYouKill.java:171) at org.eclipse.jetty.util.thread.strategy.EatWhatYouKill.run(EatWhatYouKill.java:129) at org.eclipse.jetty.util.thread.ReservedThreadExecutor$ReservedThread.run(ReservedThreadExecutor.java:388) at org.eclipse.jetty.util.thread.QueuedThreadPool.runJob(QueuedThreadPool.java:806) at org.eclipse.jetty.util.thread.QueuedThreadPool$Runner.run(QueuedThreadPool.java:938) at java.lang.Thread.run(Thread.java:748)
```
Yannick Marcon
--
You received this message because you are subscribed to the Google Groups "obiba-users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to obiba-users...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/obiba-users/6ad5bd52-a42c-4652-b1b2-f6f8f62e8a70n%40googlegroups.com.
Iago Giné Vázquez
Yannick Marcon
To view this discussion on the web visit https://groups.google.com/d/msgid/obiba-users/27ef6ae6-0145-40f2-9141-1ec82c0231e1n%40googlegroups.com.
Iago Giné Vázquez
Yannick Marcon
To view this discussion on the web visit https://groups.google.com/d/msgid/obiba-users/ae5b0c89-fee2-42c4-9f07-07eef4ac0735n%40googlegroups.com.
Iago Giné Vázquez
Iago Giné Vázquez
Cédric Fontin
Hi,
You seem to have modified the fields access.access_data, access.access_bio_samples and access.access_other study fields such that they are no longer String but rather SimpleSequence as the error suggests.
You’d have to modify the macro to take this into account.
To do so, you’d have to create a $MICA_HOME/templates/models/study.ftl on your server (use this file as an example) and write a new studyAccessInfo macro to override the default one.
You will need to restart mica afterwards (you’d need to restart everytime you add a new file).
Regards.
To view this discussion on the web visit https://groups.google.com/d/msgid/obiba-users/266544ce-2f4d-4e29-b73e-e90910df21dan%40googlegroups.com.
Cédric Fontin
To Add:
$MICA_HOME is by default /etc/mica2.
You’d also need to change the ownership of the directories and files that you add to user mica and group adm (an example command would be chown -R mica:adm $MICA_HOME (where $MICA_HOME is the true path).
Regards.
To view this discussion on the web visit https://groups.google.com/d/msgid/obiba-users/266544ce-2f4d-4e29-b73e-e90910df21dan%40googlegroups.com.
Iago Giné Vázquez
Cédric Fontin
Hi,
The change should have been done in the individual Study’ Form Configuration.
You can find that menu in the administration portal under Administration > Individual Study Configuration.
Under Form > Schema
You can search for one of the fields (e.g. access_data). It should look like this:
"access": {
"type": "object",
"format": "radioGroupCollection",
"values": [
{"key": "yes", "caption":"t(global.yes)"},
{"key": "no", "caption":"t(global.no)"},
{"key": "na", "caption":"t(global.not-applicable)"}
],
"items": [
{
"key": "access_data",
"name": "t(study_taxonomy.vocabulary.access_data.title)"
},
{
"key": "access_bio_samples",
"name": "t(study_taxonomy.vocabulary.access_bio_samples.title)"
},
{
"key": "access_other",
"name": "t(study_taxonomy.vocabulary.access_other.title)"
}
]
}
If that is not the case, you need to take note of what data structure it is and make the changes in the ftl templates.
For looping through sequences, you can check this documentation.
Regards.
To view this discussion on the web visit https://groups.google.com/d/msgid/obiba-users/806b5c38-ef27-4600-8b7e-ee1a5b04b480n%40googlegroups.com.
