DPMM with differing number of observations between subjects
35 views
Skip to first unread message
Nathan Constantine-Cooke
May 27, 2021, 10:25:31 AM5/27/21
to nimble-users
Hi all,
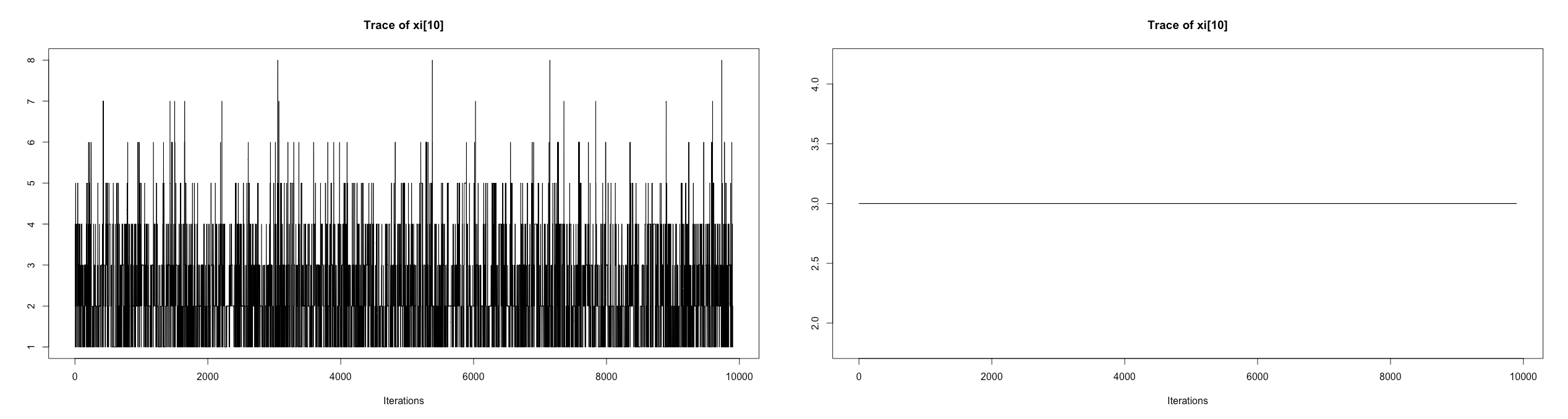
I am interested in fitting a Dirichlet Process Mixture Models clustering coefficients. I am able to fit the model and get appropriate results when the number of observations are equal between subjects (mostly matching the true clusters). However, the traces for xi do not behave as expected when the number of observations differ between subjects: seemingly becoming completely random as shown by the below plot (left: differing number of observations between subject; right: same number of observations per subject).

Am I making a mistake somewhere, or is this not currently supported by Nimble? I have tried to reduce the problem to minimum working examples based on linear regression in the attached R scripts. DPMM_MWE_working demonstrates the chains behaving correctly for equal observations between subjects, whilst DPMM_MWE_broken demonstrates the issue I have described here.
I would appreciate any help/confirmation this is a bug/confirmation this is not supported. I have checked the GitHub development branches but none seem relevant to this potential problem.
Kind regards,
Nathan
#########################################
############### DPMM_MWE_broken ##########
#########################################
## ----seed----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
set.seed(1)
library(nimble)
library(coda)
## ----basic params--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
subject.num <- 10 # Number of subjects
nmin <- 3
nmax <- 15
n <- ceiling(runif(subject.num, nmin, nmax)) # Number of measurements
maxtime <- 5
cluster_num <- 4
## ----defining true betas-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
betas <- matrix(c(rep(2, 3),
c(3.3, 2.2, -0.5),
c(1.5, 1.75, 2),
c(2.9, 1.6, 1)),
nrow = 4,
byrow = TRUE)
rownames(betas) <- paste("cluster:", seq(1, 4))
colnames(betas) <- paste0("beta_", seq(1, 3))
## ----Simulating data-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
times.mat <- matrix(nrow = 0, ncol = max(n))
sim.vals <- matrix(nrow = 0, ncol = max(n) + 2) # + 2 for subject num & true cluster
for (subject in 1:subject.num) {
clust.assign <- ceiling(cluster_num * runif(1))
times <- runif(n[subject], 0, maxtime)
times <- sort(times)
times <- c(times, rep(0, max(n) - n[subject]))
times.mat <- rbind(times.mat, times)
betas.val <- betas[clust.assign, ]
values <- rep(0, n[subject])
errors <- rnorm(n = n[subject], mean = 0, sd = sqrt(1))
for (i in seq_len(n[subject])) {
values[i] <- betas.val[1] + (times[i] * betas.val[2]) + ((times[i] ^ 2) * betas.val[3]) + errors[i]
}
values <- c(subject, clust.assign, values, rep(0, max(n) - n[subject]))
sim.vals <- rbind(sim.vals, values)
}
colnames(sim.vals) <- c("subject", "cluster", paste0("time_", seq(1, max(n))))
## ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
nburnin <- 100
thin <- 1
niter <- 10000
## ----MCMC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
subject.num <- nrow(sim.vals)
code <- nimbleCode({
# Priors
sigma2 ~ dinvgamma(shape = 1, scale = 2)
# Vector of class memberships for each subject
xi[1:subject.num] ~ dCRP(conc = alpha, size = subject.num)
#alpha ~ dgamma(shape = 1, rate = 1)
alpha ~ dinvgamma(shape = 1, scale = 1)
# betas
for (i in 1:subject.num) { # for subject i
for (j in 1:3) {
betaTilde[i, j] ~ dnorm(0, 1)
beta[i, j] <- betaTilde[xi[i], j]
}
for (t in 1:n[i]) {
logy[i, t] ~ dnorm(mean = mu[i, t], sd = sqrt(sigma2))
mu[i, t] <- beta[i, 1] + (beta[i, 2] * times[i, t]) + beta[i, 3] * (times[i, t] ^ 2)
}
}
})
constants <- list(subject.num = subject.num,
n = n)
data <- list(logy = sim.vals[, c(-1, -2)], times = times.mat)
inits <- list(beta = matrix(0, nrow = subject.num, ncol = 3),
alpha = 1,
mu <- matrix(1, nrow = subject.num, ncol = max(n)),
betaTilde = matrix(0, nrow = subject.num, ncol = 3),
xi = rep(1, subject.num),
sigma2 = 2.5)
## ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
model <- nimbleModel(code = code,
data = data,
inits = inits,
constants = constants)
model$initializeInfo()
## ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
cmodel <- compileNimble(model)
## ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
conf <- configureMCMC(model, monitors = c('xi', 'beta', 'alpha'), print = TRUE)
## ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
mcmc <- buildMCMC(conf)
cmcmc <- compileNimble(mcmc, project = model, resetFunctions = TRUE)
## ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
nimbleMCMC_samples_sim <- runMCMC(cmcmc,
nburnin = nburnin,
thin = thin,
niter = niter)
## ----Trace---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
par(mfrow = c(2, 3))
traceplot(as.mcmc(nimbleMCMC_samples_sim))
## ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
Mode <- function(x) { # Find the Mode for a vector
ux <- unique(x)
ux[which.max(tabulate(match(x, ux)))]
}
clusters <- apply(nimbleMCMC_samples_sim[, 32:41],
2,
Mode)
## ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
temp <- cbind(sim.vals[,"cluster"], clusters)
rownames(temp) <- seq(1, subject.num)
colnames(temp) <- c("true_cluster", "predicted_cluster")
temp
#########################################
############### DPMM_MWE_working #########
#########################################
## ----seed----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
set.seed(1)
library(nimble)
library(coda)
## ----basic params--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
subject.num <- 10
n <- 10
maxtime <- 5
cluster_num <- 4
## ----defining true betas-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
betas <- matrix(c(rep(2, 3),
c(3.3, 2.2, -0.5),
c(1.5, 1.75, 2),
c(2.9, 1.6, 1)),
nrow = 4,
byrow = TRUE)
rownames(betas) <- paste("cluster:", seq(1, 4))
colnames(betas) <- paste0("beta_", seq(1, 3))
## ----Simulating data-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
times.mat <- matrix(nrow = 0, ncol = n)
sim.vals <- matrix(nrow = 0, ncol = n + 2) # + 2 for subject num & true cluster
for (subject in 1:subject.num) {
clust.assign <- ceiling(cluster_num * runif(1))
times <- runif(n, 0, maxtime)
times <- sort(times)
times.mat <- rbind(times.mat, times)
betas.val <- betas[clust.assign, ]
values <- rep(0, n)
errors <- rnorm(n = n, mean = 0, sd = sqrt(1))
for (i in seq_len(n)) {
values[i] <- betas.val[1] + (times[i] * betas.val[2]) + ((times[i] ^ 2) * betas.val[3]) + errors[i]
}
values <- c(subject, clust.assign, values)
sim.vals <- rbind(sim.vals, values)
}
colnames(sim.vals) <- c("subject", "cluster", paste0("time_", seq(1, n)))
## ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
nburnin <- 100
thin <- 1
niter <- 10000
## ----MCMC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
subject.num <- nrow(sim.vals)
code <- nimbleCode({
# Priors
sigma2 ~ dinvgamma(shape = 1, scale = 2)
# Vector of class memberships for each subject
xi[1:subject.num] ~ dCRP(conc = alpha, size = subject.num)
#alpha ~ dgamma(shape = 1, rate = 1)
alpha ~ dinvgamma(shape = 1, scale = 1)
# betas
for (i in 1:subject.num) { # for subject i
for (j in 1:3) {
betaTilde[i, j] ~ dnorm(0, 1)
beta[i, j] <- betaTilde[xi[i], j]
}
for (t in 1:n) {
logy[i, t] ~ dnorm(mean = mu[i, t], sd = sqrt(sigma2))
mu[i, t] <- beta[i, 1] + (beta[i, 2] * times[i, t]) + beta[i, 3] * (times[i, t] ^ 2)
}
}
})
constants <- list(subject.num = subject.num,
n = n)
data <- list(logy = sim.vals[, c(-1, -2)], times = times.mat)
inits <- list(beta = matrix(0, nrow = subject.num, ncol = 3),
alpha = 1,
mu <- matrix(1, nrow = subject.num, ncol = max(n)),
betaTilde = matrix(0, nrow = subject.num, ncol = 3),
xi = rep(1, subject.num),
sigma2 = 2.5)
## ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
model <- nimbleModel(code = code,
data = data,
inits = inits,
constants = constants)
model$initializeInfo()
## ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
cmodel <- compileNimble(model)
## ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
conf <- configureMCMC(model, monitors = c('xi', 'beta', 'alpha'), print = TRUE)
## ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
mcmc <- buildMCMC(conf)
cmcmc <- compileNimble(mcmc, project = model, resetFunctions = TRUE)
## ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
nimbleMCMC_samples_sim <- runMCMC(cmcmc,
nburnin = nburnin,
thin = thin,
niter = niter)
## ----Trace---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
#par(mfrow = c(2, 3))
traceplot(as.mcmc(nimbleMCMC_samples_sim))
## ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
Mode <- function(x) { # Find the Mode for a vector
ux <- unique(x)
ux[which.max(tabulate(match(x, ux)))]
}
clusters <- apply(nimbleMCMC_samples_sim[, 32:41],
2,
Mode)
## ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
temp <- cbind(sim.vals[,"cluster"], clusters)
rownames(temp) <- seq(1, subject.num)
colnames(temp) <- c("true_cluster", "predicted_cluster")
temp
Claudia Wehrhahn Cortes
May 27, 2021, 3:06:07 PM5/27/21
to Nathan Constantine-Cooke, nimble-users
Hi Nathan,
thanks for using the BNP functionality in nimble!
Having a different number of observations per subject is not currently supported in nimble's CRP sampler. Internally, we are assuming that you have the same number of observations per subject. That's why you get many warnings during building and compiling.
I'm not sure if there could be a work around, maybe Chris would have a better idea. (Chris?)
BTW, in the case with the same number of observations per subject, nimble will assign a more efficient sampler for the concentration parameter if you consider a gamma prior.
Best,
Claudia
--
You received this message because you are subscribed to the Google Groups "nimble-users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to nimble-users...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/nimble-users/c3667465-ede2-4ec9-81ed-70aadf4e70f6n%40googlegroups.com.
Chris Paciorek
May 27, 2021, 5:05:19 PM5/27/21
to Claudia Wehrhahn Cortes, Nathan Constantine-Cooke, nimble-users
As we have it set up we assume the same number of observations per subject.
However, this reveals that we should be trapping this and not allowing the MCMC to be built and run. I will file an issue for us to fix this.
Nathan, a work-around would be to "fill in" the "missing" elements with NAs so that there are equal numbers per subject. The MCMC would then impute those values, which would take some extra computation, as well as potentially slowing mixing a bit. How much of an effect this would be would depend on the maximum number of observations per subject and therefore how many 'missing' elements there are if you try to fill things out so all the subjects have equal numbers of observations.
To view this discussion on the web visit https://groups.google.com/d/msgid/nimble-users/CADcEeVq9gBo752X1VW0P8znPcv4_%2B0hvLSGAnX4xMjb9y71smw%40mail.gmail.com.
Nathan Constantine-Cooke
Jun 1, 2021, 7:15:03 AM6/1/21
to nimble-users
Thank you both for your quick replies! It is good to know I've not been doing something hideously wrong with my coding.
Imputing from he MCMC seems to be working well once I made sure values were only imputed between time values I have for observed data. Thank you very much for the work around, Chris!
Kind regards,
Nathan
Reply all
Reply to author
Forward
0 new messages
