Strange traceplots–parameters not getting sampled?
Matthijs Hollanders
Matthijs Hollanders
Chris Paciorek
--
You received this message because you are subscribed to the Google Groups "nimble-users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to nimble-users...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/nimble-users/42cf8b72-966f-409b-ac01-b24b44a0dd62n%40googlegroups.com.
Matthijs Hollanders
Matthijs Hollanders
David Pleydell
Matthijs Hollanders
Jose.J...@uclm.es
Rémi Patin
I'm fairly new to custom distributions and I've recently tried to use an
hypergeometric distribution in one of my model, but I encountered an
error that I do not understand.
I cannot understand where does the error comes from, so if anyone has
any idea on how to debug this further I would be very grateful (I am
fairly confident that further bugs will appear but one step at a time).
When I run the attached script "run.R", the model is correctly loaded
and manage to run calculate().
However the first compileNimble() fails with the following error :
> Error in code[[i]] : subscript out of bounds
I attached a reproducible example with the custom distribution and
associated nimbleFunctions as well as the model and data.
To be totally transparent I based this work on this old conversation on
the list :
https://groups.google.com/g/nimble-users/c/g766aXzf5ps/m/B0NGGhAXAAAJ
I also have the secret hope that hypergeometric distribution are already
implemented in Nimble or someone has already worked with it, so feel
free to point out such information :-)
Cheers,
Rémi
Daniel Turek
run = function(n = double(0), k = double(0)){
returnType(double(0))
# if(any(is.na(c(n,k)))) return(NA)
if(k != 0) {
n <- round(n)
k <- round(k)
return(sum(log((n-k+1):n)) - sum(log(1:k)))
} else {
return(0)
}
}
)
--
You received this message because you are subscribed to the Google Groups "nimble-users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to nimble-users...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/nimble-users/bc38c4c1-c0e2-98cd-b9fa-d415a5747774%40gmail.com.
Perry de Valpine
To view this discussion on the web visit https://groups.google.com/d/msgid/nimble-users/CAKbe0ho57FpiOiu0FtNJqTO2y_r7M_MfKcFL-owTfkzcv19PFg%40mail.gmail.com.
Chris Paciorek
To view this discussion on the web visit https://groups.google.com/d/msgid/nimble-users/a04c6479-7a64-4c3a-9d49-d3205990a294n%40googlegroups.com.
Jose Jimenez Garcia-Herrera
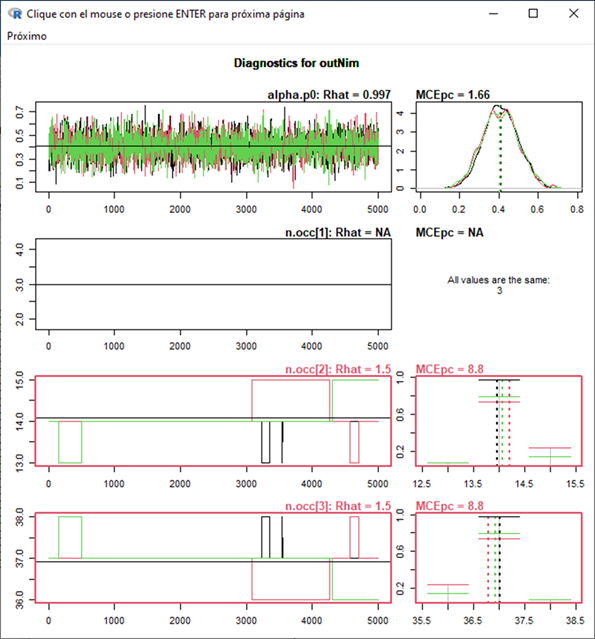
Hi Chris. Thanks for the response. In a multi-state model (with many NA data) the sampler seems not to update the occupancy values for the different states, with many of the z posteriors "frozen".
……
# DERIVED QUANTITIES
#####################
for (i in 1:M){
occ1[i] <- equals(z[i], 1)
occ2[i] <- equals(z[i], 2)
occ3[i] <- equals(z[i], 3)
}
n.occ[1] <- sum(occ1[1:M]) # Sites in state 1
n.occ[2] <- sum(occ2[1:M]) # Sites in state 2
n.occ[3] <- sum(occ3[1:M]) # Sites in state 3
………
z Posteriors:
z[1] 3.00000 0.000000 0.000e+00 0.0000000
z[2] 3.00000 0.000000 0.000e+00 0.0000000
z[3] 3.00000 0.000000 0.000e+00 0.0000000
z[4] 3.00000 0.000000 0.000e+00 0.0000000
z[5] 3.00000 0.000000 0.000e+00 0.0000000
z[6] 3.00000 0.000000 0.000e+00 0.0000000
z[7] 3.00000 0.000000 0.000e+00 0.0000000
z[8] 3.00000 0.000000 0.000e+00 0.0000000
z[9] 3.00000 0.000000 0.000e+00 0.0000000
z[10] 2.00000 0.000000 0.000e+00 0.0000000
z[11] 2.90307 0.295877 2.416e-03 0.1091225
z[12] 2.58967 0.491911 4.016e-03 0.1263183
z[13] 2.00000 0.000000 0.000e+00 0.0000000
z[14] 2.00000 0.000000 0.000e+00 0.0000000
z[15] 3.00000 0.000000 0.000e+00 0.0000000
z[16] 2.02847 0.166307 1.358e-03 0.0383330
z[17] 3.00000 0.000000 0.000e+00 0.0000000
z[18] 2.82540 0.379637 3.100e-03 0.1412142
Chris Paciorek
Jose Jimenez Garcia-Herrera
Thank you Chris for your answer. Attached you can find the code and a subset of the data. I am aware of the amount of NA, but it works in WinBUGS and JAGS. That is why it was my question about the sampler.
Best regards,
Jose
Chris Paciorek
Jose Jimenez Garcia-Herrera
Thank you, Chris. Your explanations make perfect sense. I noticed that as missing data increases, correct sampling becomes more difficult. Using other inits values on z is not so easy, because I sent you a simplified code. Real code is extremely complex... But to clarify, your proposal is rewriting the MS code to avoid having to update the missing values and supply only the non-missing values as data. Isn’t it?
Best regards
To view this discussion on the web visit https://groups.google.com/d/msgid/nimble-users/CAG%3DM9LqM1WvdvjxHBMf-G_zo_pFh0oAHHh6PCVGw%2B%2BATu6JEcA%40mail.gmail.com.
Chris Paciorek
David Christianson
Chris Paciorek
To view this discussion on the web visit https://groups.google.com/d/msgid/nimble-users/eb2c9b57-a504-40c4-8b11-0f06369e45a0n%40googlegroups.com.
