Sampling (out of sample) from posterior predictive distribution
240 views
Skip to first unread message
Lucas Godoy
Aug 9, 2022, 4:11:02 PM8/9/22
to nimble-users
Hello everyone,
I have a geostatistical model and want to sample from the posterior predictive distribution at a new set of locations. So far, the only way I could do it was by setting an "augmented" model with NAs at the locations at which I want to sample from this posterior predictive distribution. However, this approach is not ideal for (at least) two reasons:
1) it affects the mixing of the chains
2) what if I don't know the regions where I want these new samples a priori?
Are there any other ways of doing it?
An option, if possible, would be to increase the dimension of some nodes after running the MCMC and then simulate the data from the model. I don't know how to do it though.
I have looked into the documentation and the email list and have not found anything helpful in this regard yet.
Any help or resources are very much appreciated.
P.s.: I have attached a (simplified) reproducible toy example to make what I'm trying to say clearer.
Thank you in advance.
Best,
Lucas.
Chris Paciorek
Aug 9, 2022, 7:58:33 PM8/9/22
to Lucas Godoy, nimble-users
Hi Lucas,
Right, you generally don't want to add the predictive nodes into the model for the reasons you say. We have an open to-do item to detect such predictive nodes and have their sampling be done only in 'predictive mode' without influencing the posterior sampling of everything else, but it's not something we've been able to put time into yet.
We have a simple example of posterior predictive sampling here that shows simulation into nodes that already exist in the model.
To do what you want, one approach would be:
1) Run the MCMC on the original model (using only the observed locations)
2) Set up a 'full' model that includes declarations for the spatial process at the original locations and for the spatial process at the new locations conditional on the original locations. You'll need to code up the conditional multivariate normal calculations for your declaration for the new locations. (You could use definition-time if-then-else to avoid having to write the model code twice, but that's a minor thing.)
3) Use the ideas from #1 to put the sampled values (process and relevant hyperparameters) into the full model and simulate process values at the new locations conditional on the posterior samples.
Happy to iterate if you have questions or get stuck.
-chris
--
You received this message because you are subscribed to the Google Groups "nimble-users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to nimble-users...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/nimble-users/6ddfb2cb-3351-4a03-90c6-c12fa1d47317n%40googlegroups.com.
Lucas Godoy
Aug 12, 2022, 8:25:32 PM8/12/22
to paci...@stat.berkeley.edu, nimble-users
Hey Chris,
Firstly, thanks for your help and prompt response!
At first sight, I thought it had worked, however, I'm not sure.
Here go some thoughts:
1) the if-then-else approach has not worked. The reason is that is says the "cov_pred" matrix is not positive definite when trying to calculate its Cholesky decomposition. I believe it might be due to numerical problems.
2) the way I created the function to calculate the predictions we pass a "target" node and the function uses `model$getParents` to track the parents of the target node. What I notice is that the grand-parents of the node are not being tracked by that function. For instance, "lifted" nodes needed to simulate `s_pred` (which is needed to predict the new data points) do not appear when I call `model$getParents` at the positions of "y" to which I want to calculate the predictions. Would "simulate" automatically deal with those lifted nodes?
3) when I plotted the predicted (posterior mean of the predictive distribution at each point t) vs the true it does not look right.
Any thoughts?
Also, is there a "chol2inv" function in Nimble?
Thank you agagin.
Best,
Lucas.
Chris Paciorek
Aug 16, 2022, 1:32:44 PM8/16/22
to Lucas Godoy, nimble-users
Hi Lucas,
1) The positive-definiteness is a numerical issue that should be unrelated to getting if-then-else to work. It works if you just define the predictive model separately?
2) No, simulate won't automatically deal with the graph structure. You need to make sure that you are telling simulate exactly what needs to be simulated, including all lifted nodes. I think what you need is "getParents("s_pred[1:10]", determOnly = TRUE)' and then concatenate "s_pred[1:10]" onto that, since with determOnly, it won't include 's_pred' since it is stoch. All the stochastic parents(phi, tau, s) should be filled with values from the original MCMC. ("10" here is just a placeholder...)
3) This likely relates at least in part to #2. It looks like you might be simulating 's' in the predictive sampling when you should be conditioning on its values from the initial MCMC.
4) No, there's not a chol2inv function, though you could implement the equivalent using chol(), forwardsolve() and backsolve().
Side note: your `code_pred` doesn't need to include 'y' since it's not involved in the predictive simulation of s_pred.
-chris
Lucas Godoy
Aug 19, 2022, 11:58:46 AM8/19/22
to paci...@stat.berkeley.edu, nimble-users
Hi Chris,
1) Yes. That was weird, probably something wrong with my code though.
2) Thanks, that makes a lot of sense.
3) About that, when I run configureMCMC(model_pred) I get the following output
===== Monitors =====
thin = 1: beta, phi, tau
===== Samplers =====
posterior_predictive_branch sampler (3)
- phi
- tau
- beta
thin = 1: beta, phi, tau
===== Samplers =====
posterior_predictive_branch sampler (3)
- phi
- tau
- beta
shouldn't "s" and "s_pred" be there too? What if I include RW block samplers for these latent variables?
I believe that will not change anything, though.
Also, I'm more interested in simulating from the posterior predictive of "y" at the new locations instead of "s_pred".
I end up getting results with the attached code, but they don't seem quite right.
Thanks again for your help.
Lucas.
Chris Paciorek
Aug 19, 2022, 1:07:31 PM8/19/22
to Lucas Godoy, nimble-users
Hi Lucas,
Big picture -- you don't need to set up an MCMC for the predictive model. You just need to use `simulate` on the appropriate nodes in `ppSamplerNF`, including any y's that you want to sample from the predictive distribution.
That being said, you could run the MCMC to do the simulation as well, so long as you make sure the only samplers are the predictive samplers that sample the nodes of interest. But in that case you'd still need to make sure that any deterministic parents have their values properly calculated given the values you put into the parameters from the original MCMC, before running the predictive samplers, so I don't think there is any benefit to do that rather than just using `simulate`.
As far as why `s` and `y` don't appear, the 'branch' in 'posterior_predictive_branch' relates to the fact that such samplers sample the target node and all downstream predictive nodes as a group, since there are not downstream data nodes. It's just that the downstream nodes that are sampled are not indicated in the information printed out. E.g., consider this model: y ~ dnorm(theta, 1); theta ~ dnorm(0,1). If you don't set 'y' as data, then the whole thing is predictive and a posterior_predictive_branch sampler would be assigned to theta and would also include conditional sampling of y given the sampled value of theta.
I haven't looked in detail at your code to see why you're getting weird results, but hopefully the above helps you figure it out.
-chris
Lucas Godoy
Aug 27, 2022, 1:30:22 PM8/27/22
to paci...@stat.berkeley.edu, nimble-users
Hey Chris,
Thank you for taking so much time to explain this in detail.
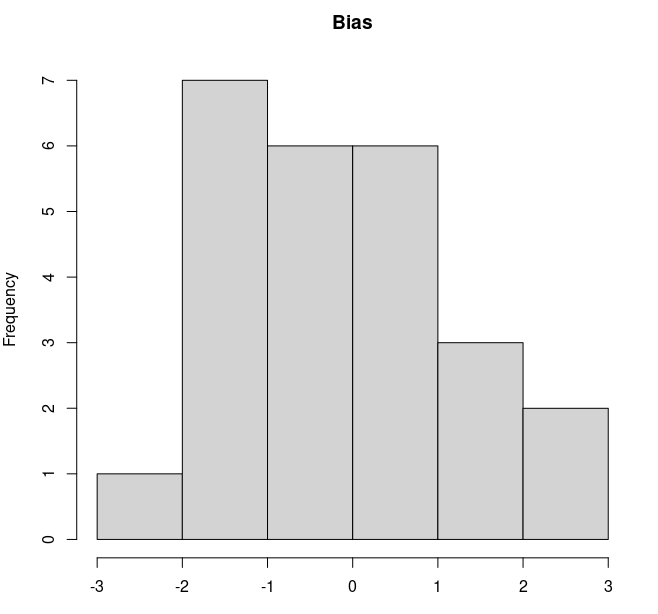
It ended up working fine, the bias of the predicted values (only 25 data points) is somewhat scattered around zero as expected.
I am attaching the code in case someone is interested.
Thanks,
Lucas.
Reply all
Reply to author
Forward
0 new messages
