How to use NGLess to generate gene count matrix?
41 views
Skip to first unread message
Yu-Jen Chang
Sep 28, 2021, 1:12:18 AM9/28/21
to NGLess
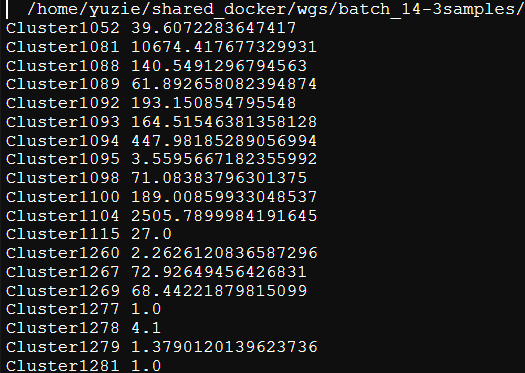
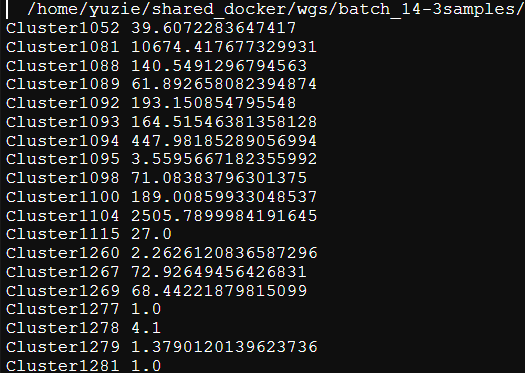
I used your tool NGLess to generate count table by using your human-gut.ngl profiler, but the output is doesn't meet my need.
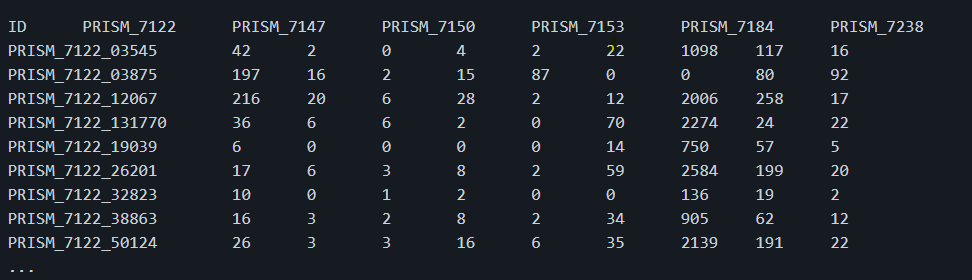

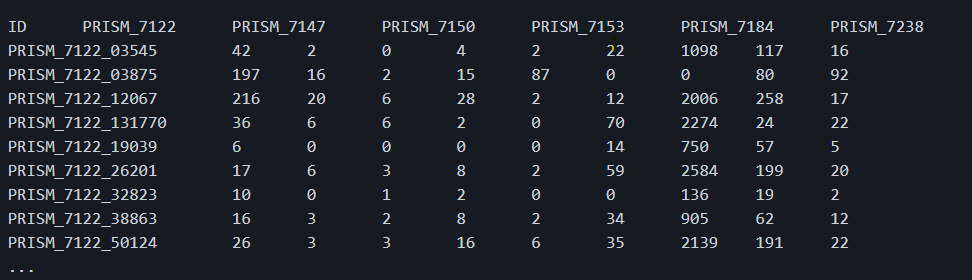
I want to generate a tab-separated values matrix giving the number of reads mapped on genes (rows) across metagenomic samples (columns).

I am not familiar with metagenomic WGS pipeline, so I really need your help !!
Thank you!
All the best,
Yu-Jen
Luis Pedro Coelho
Oct 1, 2021, 5:04:18 AM10/1/21
to NGLess List
Dear Yu-Jen,
(Sorry for the delay in replying, this was a busy week).
The way to approach this within NGLess is to use the parallel module:
Also, do note that the motus version from that script is now a bit outdated. Because of a desire to keep backwards compat, I have hesitated a bit too long to update the ng-meta-profiler repository, but I just did so
HTH,
Luis
Luis Pedro Coelho | Fudan University | http://luispedro.org
--You received this message because you are subscribed to the Google Groups "NGLess" group.To unsubscribe from this group and stop receiving emails from it, send an email to ngless+un...@googlegroups.com.To view this discussion on the web visit https://groups.google.com/d/msgid/ngless/cd350b39-877a-4240-8c89-77b796e1746dn%40googlegroups.com.
Reply all
Reply to author
Forward
0 new messages