Cells' consumption of cytokine concentration
Victoria Ponce
Walter de Back
A cell can sense and consume the cytokine concentration using a Mapper as in https://morpheus.gitlab.io/blog/2018/02/13/a-multiscale-mini-model/However how can I decrease the concentration of the cytokine after the cell has consumed it?
Good question, this is not obvious to many new users.
The logic is like this: In your PDE that describes the reaction and diffusion of your cytokine, you include a negative term (i.e. for decrease) that is nonzero where there is a cell and zero where there is no cell.
The attached model contains minimal example, resulting in this situation:
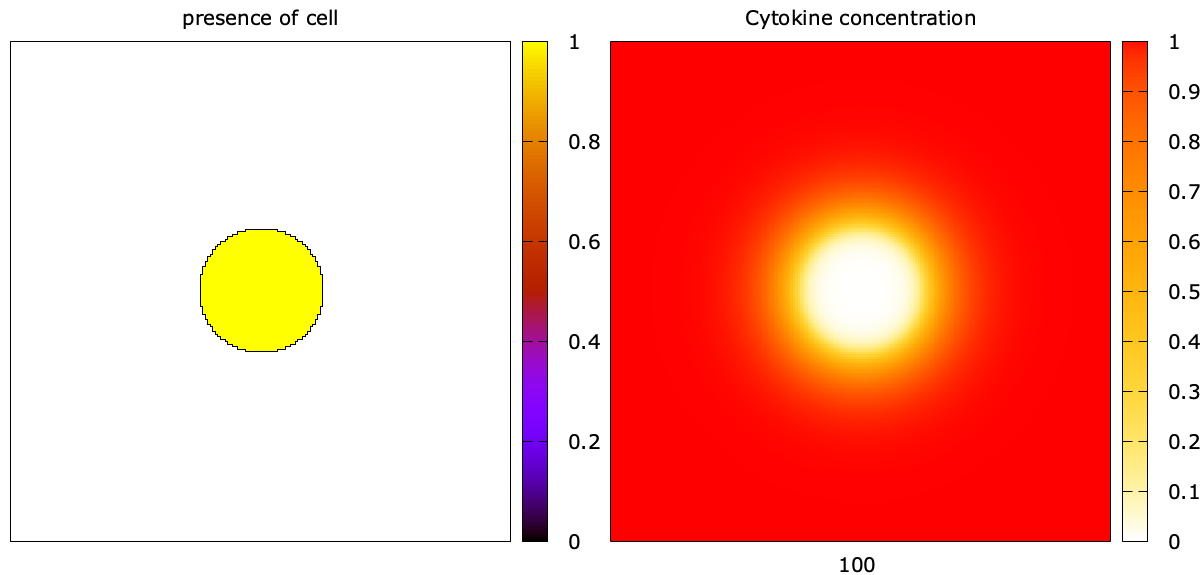
First, in Global, I defined a Field called cytokine with an initial concentration of 1.0 and diffusion.
<Field symbol="cytokine" value="1.0" name="Cytokine concentration">
<Diffusion rate="1"/>
</Field>
Then, I defined a System with a differential equation with a negative term -r*cell*cytokine where r is the consumption rate of cells. (Here r is a constant, but it could also be a cell property to specify that each cell has its own consumption rate.)
<System solver="adaptive45">
<Constant symbol="r" value="0.1" name="consumption rate"/>
<DiffEqn symbol-ref="cytokine">
<Expression>
-r*cell*cytokine
</Expression>
</DiffEqn>
</System>
Finally, I defined a Constant called cell which is 0 everywhere, except where there is a cell.
To do this, specify cell in Global and set it to 0:
<Global>
<Constant symbol="cell" value="0" name="presence of cell"/>
</Global>
But in CellType, the value of the constant cell is locally overwritten with a value of 1:
<CellType class="biological" name="cells">
<Constant symbol="cell" value="1" name="presence of cell"/>
</CellType>
In this way, the term "-r*cell*cytokine" is negative 'within' cells and zero outside. Therefore, the cytokine concentration is only decreased at locations with cells.
See the attached model for more details. Hope this helps!
