Saving minian output as a matlab file
468 views
Skip to first unread message
ph.va...@gmail.com
Jan 7, 2022, 10:37:00 AM1/7/22
to Miniscope
Hi all,
I used the minian pipeline to analyse one of my sessions and I am really happy with the results (based on the final viaualisation), so first of all - big thank you to the developer(s) for sharing this tool with the community!
This is, however, more or less the first time I use python and I am a bit lost on how to convert the output to a Matlab file. What I want to get is three matlab matrices containing the A, C and S data from the pipeline.
I found this in the docs: https://minian.readthedocs.io/en/stable/tips/variables.html#saving-to-other-formats
However, when I try to run:
minian_ds = open_minian(dpath)
I get:
GroupNotFoundError: group not found at path ''
I tried:
minian_ds = open_minian(dpath + '/minian')
And I got a minian_ds var of type xarray.core.dataset.Dataset
However, I am a bit lost as to how to proceed. The final line in the docs says run
minian_ds["C"].values.save("C.npy") to get the npy and then convert to matlab but it gives me
AttributeError: 'numpy.ndarray' object has no attribute 'save'
I hope it's as simple as I think it is and someone could lend a hand!
Best,
PV
Federico Sangiuliano
Jan 7, 2022, 10:17:43 PM1/7/22
to Miniscope
Hello PV. One way of loading the output from Minian in MATLAB is to save the output in the netCDF format. To achieve this, at the bottom of the Jupyter notebook you have to add a cell with the following code
This should save you a file named "minian_dataset" on the same folder where the output videos are located. Then, in MATLAB you can load the data like this
cd YOUR_PATH_TO_minian_dataset.nc
Sdata = ncread('minian_dataset.nc','S');
As an example, I processed three videos from my own recordings with the v4 Miniscope. Each video has 1000 frames, so in total I processed 3000 frames with a size of 608x608 pixels. It's easy to check this information from the notebook output of "varr"
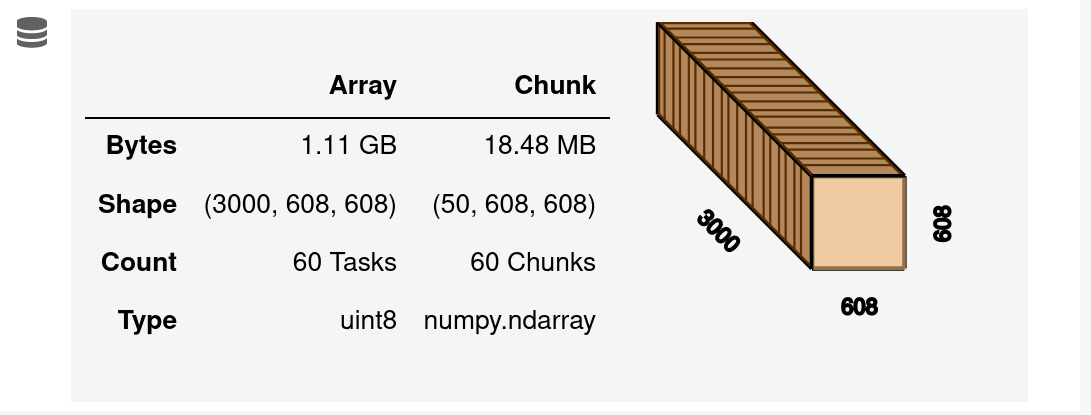
By processing my data I got 606 neurons. Adata will hold the footprint of each neuron. Running "whos Adata" will give the following information
Name Size Bytes Class Attributes
Adata 608x608x606 1792131072 double
Adata 608x608x606 1792131072 double
The structure of Adata is Adata(x,y,cell_number) where x and y will go from 1 to 608 and cell_number from 1 to 606. For C, the output from "whos Cdata" results in
Name Size Bytes Class Attributes
Cdata 3000x606 14544000 double
Cdata 3000x606 14544000 double
The structure of Cdata is Cdata(amplitude_frame,cell_number), where amplitude_frame will be the amplitude of C in that frame for a given cell_number.
I hope this helps!
Federico
Philip Vassilev
Jan 8, 2022, 5:09:14 AM1/8/22
to Federico Sangiuliano, Miniscope
Thank you, Federico, worked like a charm, it's exactly what I was looking for!
Kind regards,
PV
--
You received this message because you are subscribed to a topic in the Google Groups "Miniscope" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/miniscope/fW7xGqWLd4E/unsubscribe.
To unsubscribe from this group and all its topics, send an email to miniscope+...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/miniscope/bcf68dff-b93f-47b8-8755-45eeabf32adfn%40googlegroups.com.
Mark Sanders
Mar 25, 2022, 6:21:00 PM3/25/22
to Miniscope
function MinianNetCDFtoMatlab()
%Grazie mille Federico Sangiuliano!
%% SELECT FOLDER WITH DATA
%folder = ['C:\Users\huberlab\Desktop\attempt 3'];
NCDFfolder = 'C:\Users\huberlab\Desktop\Bruno rig tests\Vibration_test_bruno_rig_1\ZB_81\customEntValHere\2022_03_10\20_59_58\My_V4_Miniscope\analsis attempts\4';
%look at what is contained in your data!
ncdisp([NCDFfolder '\' 'minian_dataset.nc']);
%The most usefull data will be saved in the following code
%see below which variables are not extracted from the .nc file
%% Read in tha data as Matlab variables
% files with xdata include all units, even those excluded in
% minian.visualization.CNMFViewer(), see label
%% Saves it to a .mat file in the same folder
save([NCDFfolder '\' 'netCDFData.mat'])
end
%Grazie mille Federico Sangiuliano!
%% SELECT FOLDER WITH DATA
%folder = ['C:\Users\huberlab\Desktop\attempt 3'];
NCDFfolder = 'C:\Users\huberlab\Desktop\Bruno rig tests\Vibration_test_bruno_rig_1\ZB_81\customEntValHere\2022_03_10\20_59_58\My_V4_Miniscope\analsis attempts\4';
%look at what is contained in your data!
ncdisp([NCDFfolder '\' 'minian_dataset.nc']);
%The most usefull data will be saved in the following code
%see below which variables are not extracted from the .nc file
%% Read in tha data as Matlab variables
% files with xdata include all units, even those excluded in
% minian.visualization.CNMFViewer(), see label
Adata = ncread([NCDFfolder '\' 'minian_dataset.nc'],'A');
Cdata = ncread([NCDFfolder '\' 'minian_dataset.nc'],'C');
Sdata = ncread([NCDFfolder '\' 'minian_dataset.nc'],'S');
c0data = ncread([NCDFfolder '\' 'minian_dataset.nc'],'c0');
b = ncread([NCDFfolder '\' 'minian_dataset.nc'],'b');
b0 = ncread([NCDFfolder '\' 'minian_dataset.nc'],'b0');
f = ncread([NCDFfolder '\' 'minian_dataset.nc'],'f');
YrAdata = ncread([NCDFfolder '\' 'minian_dataset.nc'],'YrA');
units = ncread([NCDFfolder '\' 'minian_dataset.nc'],'unit_id');
unit_labels = ncread([NCDFfolder '\' 'minian_dataset.nc'],'unit_labels');
max_proj = ncread([NCDFfolder '\' 'minian_dataset.nc'],'max_proj');
%Less usefull variables: height, width, frame, motion
% height = ncread([NCDFfolder '\' 'minian_dataset.nc'],'height');
% width = ncread([NCDFfolder '\' 'minian_dataset.nc'],'width');
% motion = ncread([NCDFfolder '\' 'minian_dataset.nc'],'motion');
%Variables; animal, session, shift_dim are of an unsupported datatype and
% cannot be extracted with ncread
%% Make version of variables with bad cells (label =-1) removed
indx = 0 <= unit_labels;
ids = unit_labels(indx);
A = Adata(:,:,indx);
C = Cdata(:,indx);
S = Sdata(:,indx);
c0 = c0data(:,indx);
YrA = YrAdata(:,indx);
clear indx
Cdata = ncread([NCDFfolder '\' 'minian_dataset.nc'],'C');
Sdata = ncread([NCDFfolder '\' 'minian_dataset.nc'],'S');
c0data = ncread([NCDFfolder '\' 'minian_dataset.nc'],'c0');
b = ncread([NCDFfolder '\' 'minian_dataset.nc'],'b');
b0 = ncread([NCDFfolder '\' 'minian_dataset.nc'],'b0');
f = ncread([NCDFfolder '\' 'minian_dataset.nc'],'f');
YrAdata = ncread([NCDFfolder '\' 'minian_dataset.nc'],'YrA');
units = ncread([NCDFfolder '\' 'minian_dataset.nc'],'unit_id');
unit_labels = ncread([NCDFfolder '\' 'minian_dataset.nc'],'unit_labels');
max_proj = ncread([NCDFfolder '\' 'minian_dataset.nc'],'max_proj');
%Less usefull variables: height, width, frame, motion
% height = ncread([NCDFfolder '\' 'minian_dataset.nc'],'height');
% width = ncread([NCDFfolder '\' 'minian_dataset.nc'],'width');
% motion = ncread([NCDFfolder '\' 'minian_dataset.nc'],'motion');
%Variables; animal, session, shift_dim are of an unsupported datatype and
% cannot be extracted with ncread
%% Make version of variables with bad cells (label =-1) removed
indx = 0 <= unit_labels;
ids = unit_labels(indx);
A = Adata(:,:,indx);
C = Cdata(:,indx);
S = Sdata(:,indx);
c0 = c0data(:,indx);
YrA = YrAdata(:,indx);
clear indx
%% Saves it to a .mat file in the same folder
save([NCDFfolder '\' 'netCDFData.mat'])
end
Op zaterdag 8 januari 2022 om 11:09:14 UTC+1 schreef ph.va...@gmail.com:
Reply all
Reply to author
Forward
0 new messages
