Thy1-GCamp6 cortical imaging with miniscope V4
fro...@gmail.com
Hi all!
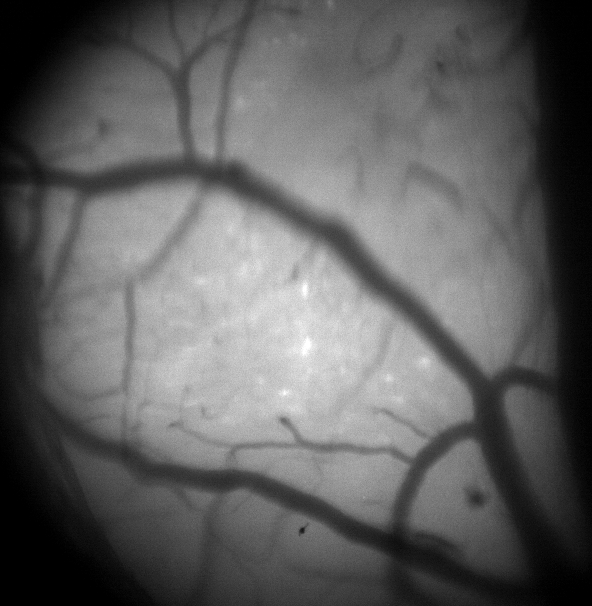
We are trying to use the Miniscope V4 with Thy1-GCaMP6sGP4.3 transgenic mice. We tried to image from the visual cortex of 2 different mice over a 4mm cranial window with a glass coverslip. We can clearly see the vessels at the top of the cortex, and we have tried to image at different depths from L1 to L2/3. However, we can never see cells in either the raw image or the processed df/f denoised image. There is calcium activity though and when we use the Minian (Dong et al. 2021) processing package, there algorithm can separate some cells. These are never clearly seen in the images (example attached).
We are wondering if this could be because of the mouse line and cortical imaging (such as dense expression of GCamp6 in neuropil) or if is a miniscope issue.
Any suggestions would be greatly appreciated. Thank you in advance!
Cheers,
Emmanouil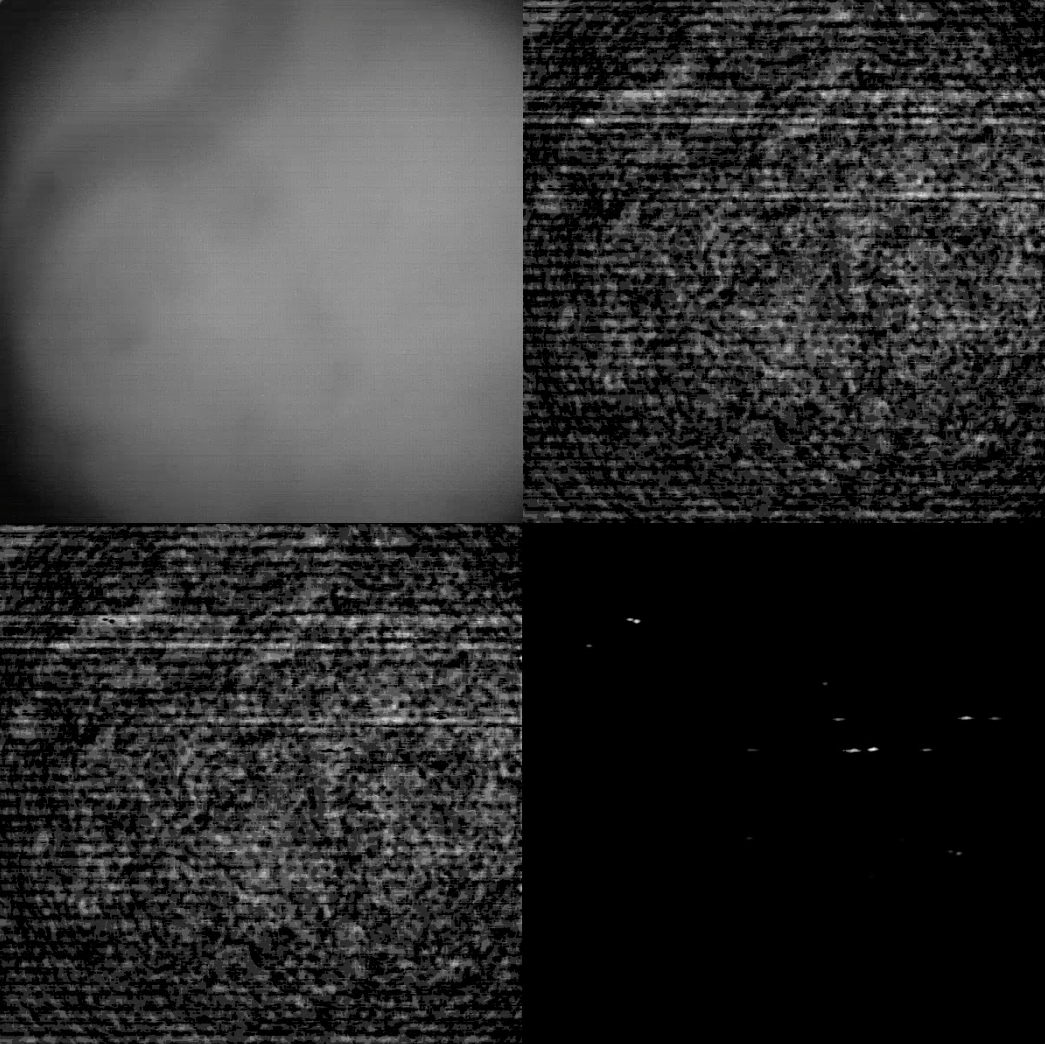
Federico Sangiuliano
fro...@gmail.com
Federico Sangiuliano
fro...@gmail.com
Alan
Daniel Aharoni
- Is this with the Thy1-GCaMP6sGP4.3?
- So you think we were too deep and should get closer to the surface as you are?
- Also your line noise levels are much lower. How long is your cable between the miniscope and the DAQ?
