Wrapped coordinates and kd-tree with PBC
124 views
Skip to first unread message
Thibaut
Sep 2, 2022, 5:28:28 AM9/2/22
to MDnalysis discussion
Hello,
I have a problem in generating a kdtree using wrapped (by MDA) coordinates.
If I use something like :
u = mda.Universe(data,dump)
boxsize = u.dimensions[:3]
hydrogens = u.select_atoms("type 5")
# Wrap coord
ag = u.atoms
transform = transformations.wrap(ag)
u.trajectory.add_transformations(transform)
for i, ts in enumerate(u.trajectory):
tree_h = KDTree(hydrogens.positions, boxsize=boxsize[:3])
It results in "ValueError: Some input data are greater than the size of the periodic box."
Indeed, boxsize[:3] = array([18.68471909, 18.68471909, 18.68471909])
And hydrogens.positions.min() = 0.0018157959
hydrogens.positions.max() = 18.684723
Do you have an idea on how to fix this? My trajectory was generated in NVT ensemble. I tried boxsize = u.dimensions.astype(np.float64) without success.
Thanks !
Oliver Beckstein
Sep 2, 2022, 1:52:46 PM9/2/22
to mdnalysis-discussion
Hi Thibaut,
What box shape are you using? Why don’t you pass all of u.dimensions to the box argument?
Presumably, the NVT simulation maintains the box. Did you check that this is true?
Why not pass the current dimensions for each step
KDTree(…, box=ts.dimensions)
(ts.dimensions is the same as u.dimensions, I just like to indicate that it’s really a property of the tlmestep).
Is KDTree our PeriodicKDTree https://docs.mdanalysis.org/stable/documentation_pages/lib/pkdtree.html or something else?
It’s still possible that there are rounding issues somewhere — wrapping is tricky business and prone to all kinds of precision problems but we can think about this when we have excluded other potential issues.
Oliver
--
You received this message because you are subscribed to the Google Groups "MDnalysis discussion" group.
To unsubscribe from this group and stop receiving emails from it, send an email to mdnalysis-discus...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/mdnalysis-discussion/2c34cf87-0c4c-4454-bb56-e79c8fb0248cn%40googlegroups.com.
--
Oliver Beckstein (he/his/him)
GitHub: @orbeckst
MDAnalysis – a NumFOCUS fiscally sponsored project
Thibaut
Nov 4, 2022, 10:01:47 AM11/4/22
to MDnalysis discussion
Hello Oliver, thank you for your answer, and sorry for the delay, I did not receive any email warning me about your answer.
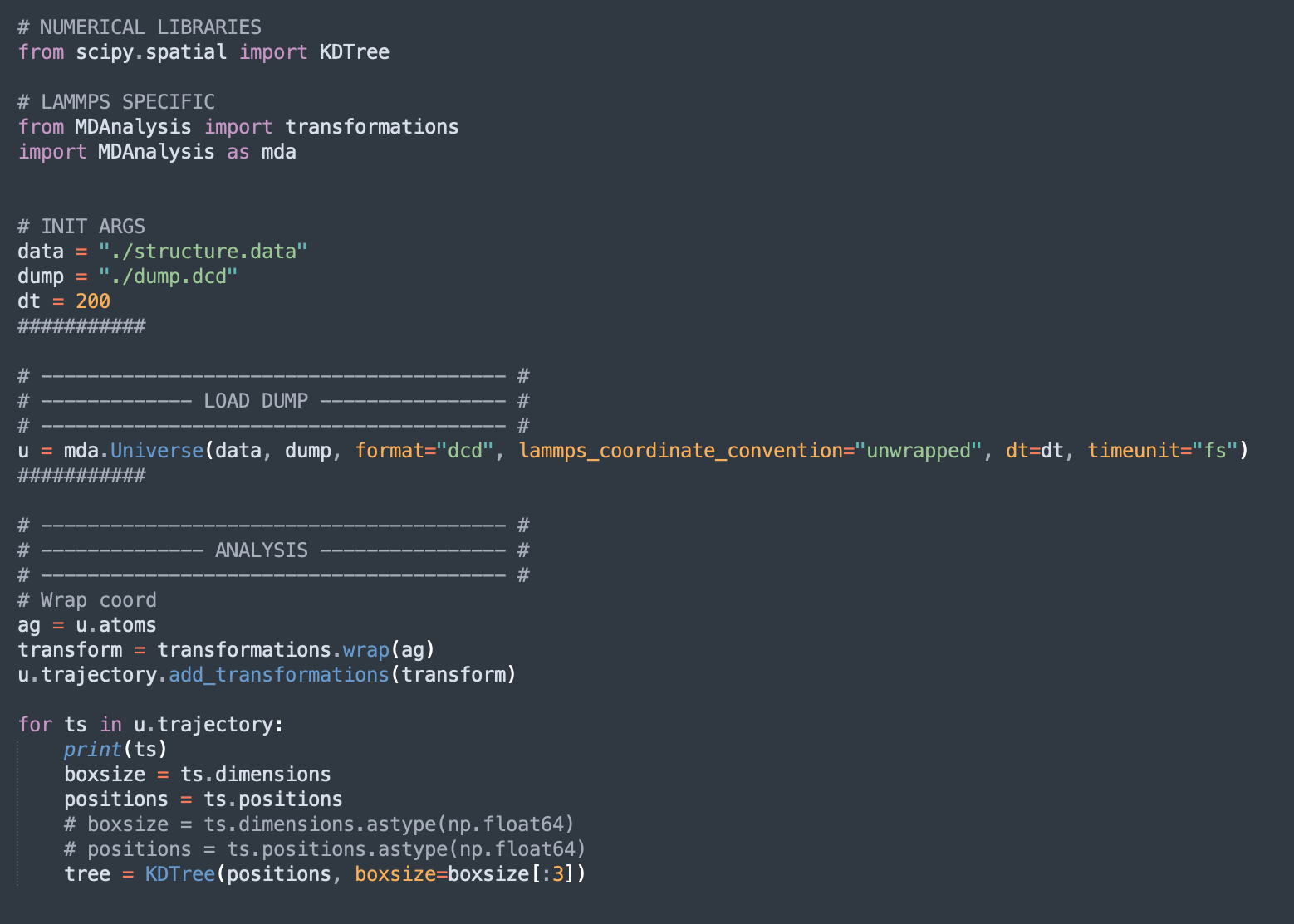
# Wrap coord
ag = u.atoms
transform = transformations.wrap(ag)
u.trajectory.add_transformations(transform)
About my problem :
- it is a cubic box
- it is a NVT run, box size is conserved
- I am using KDTree from scipy
- scipy KDTree only use cubic boxes to handle PBC, and take an array of shape (3,)
Additional infos :
- it is a dcd lammps trajectory.
Here is a minimal example :

```
# NUMERICAL LIBRARIES
from scipy.spatial import KDTree
# LAMMPS SPECIFIC
from MDAnalysis import transformations
import MDAnalysis as mda
# INIT ARGS
data = "./structure.data"
dump = "./dump.dcd"
dt = 200
###########
# ---------------------------------------- #
# ------------- LOAD DUMP ---------------- #
# ---------------------------------------- #
u = mda.Universe(data, dump, format="dcd", lammps_coordinate_convention="unwrapped", dt=dt, timeunit="fs")
###########
# ---------------------------------------- #
# -------------- ANALYSIS ---------------- #
# ---------------------------------------- #
from scipy.spatial import KDTree
# LAMMPS SPECIFIC
from MDAnalysis import transformations
import MDAnalysis as mda
# INIT ARGS
data = "./structure.data"
dump = "./dump.dcd"
dt = 200
###########
# ---------------------------------------- #
# ------------- LOAD DUMP ---------------- #
# ---------------------------------------- #
u = mda.Universe(data, dump, format="dcd", lammps_coordinate_convention="unwrapped", dt=dt, timeunit="fs")
###########
# ---------------------------------------- #
# -------------- ANALYSIS ---------------- #
# ---------------------------------------- #
# Wrap coord
ag = u.atoms
transform = transformations.wrap(ag)
u.trajectory.add_transformations(transform)
for ts in u.trajectory:
print(ts)
boxsize = ts.dimensions
positions = ts.positions
# boxsize = ts.dimensions.astype(np.float64)
# positions = ts.positions.astype(np.float64)
tree = KDTree(positions, boxsize=boxsize[:3])
```
print(ts)
boxsize = ts.dimensions
positions = ts.positions
# boxsize = ts.dimensions.astype(np.float64)
# positions = ts.positions.astype(np.float64)
tree = KDTree(positions, boxsize=boxsize[:3])
```
I am sure that it is a "problem" with wrap function, as when I get a "ValueError: Some input data are greater than the size of the periodic box.", the maximum position is beyond the box limit (positions[:,2].max() > box size[0]). I tried to play with precisions without success. Maybe I should add a translation to push everything in the box ? However, I would like to be sure that wrap function is reliable.
I can share dump.dcd and structure.data files, but I am not sure how dcd files are portables.
Reply all
Reply to author
Forward
0 new messages
