[maker-devel] cds error
70 views
Skip to first unread message
zc y
Dec 17, 2021, 12:16:03 PM12/17/21
to maker...@yandell-lab.org
Dear Maker developers,
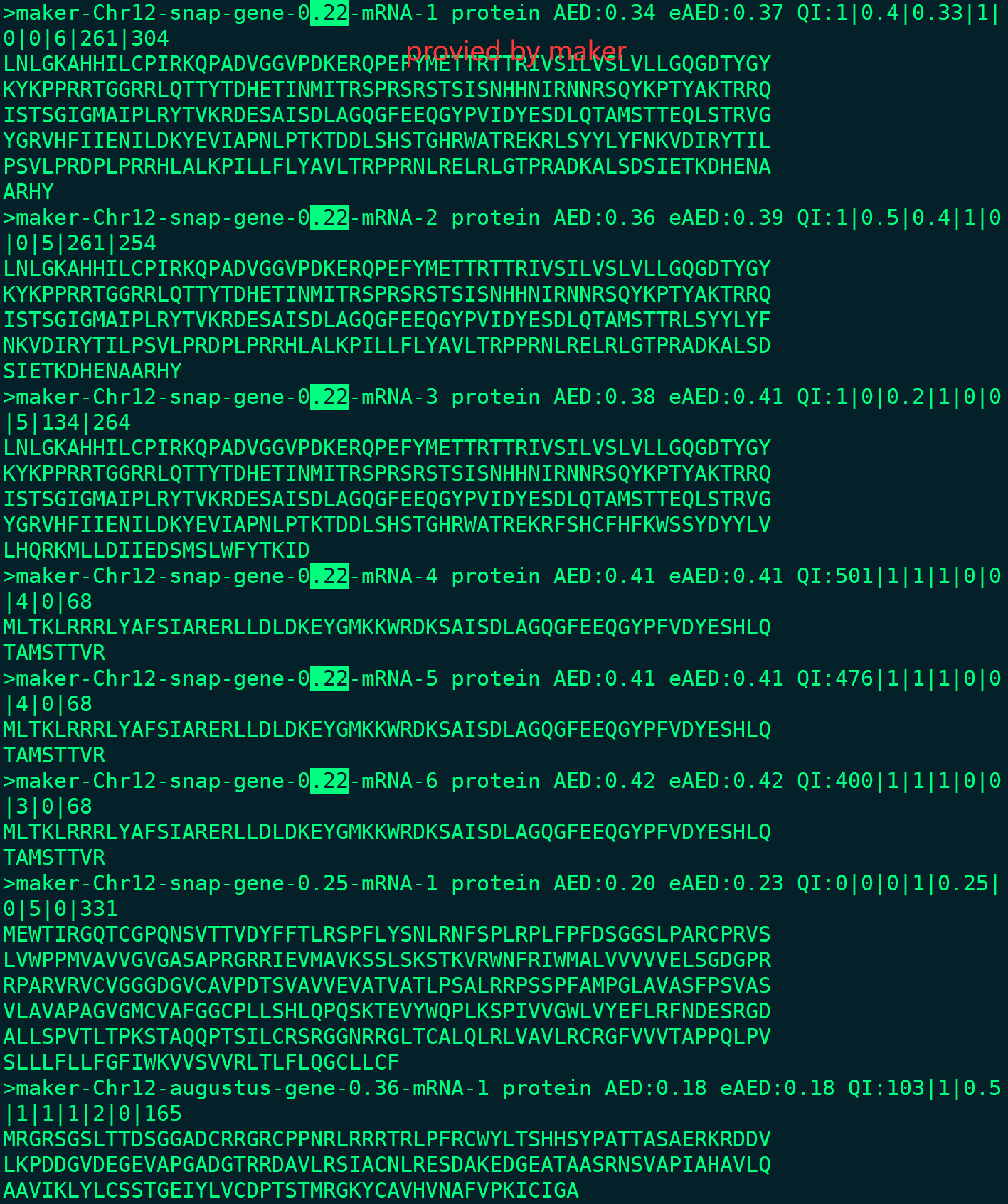
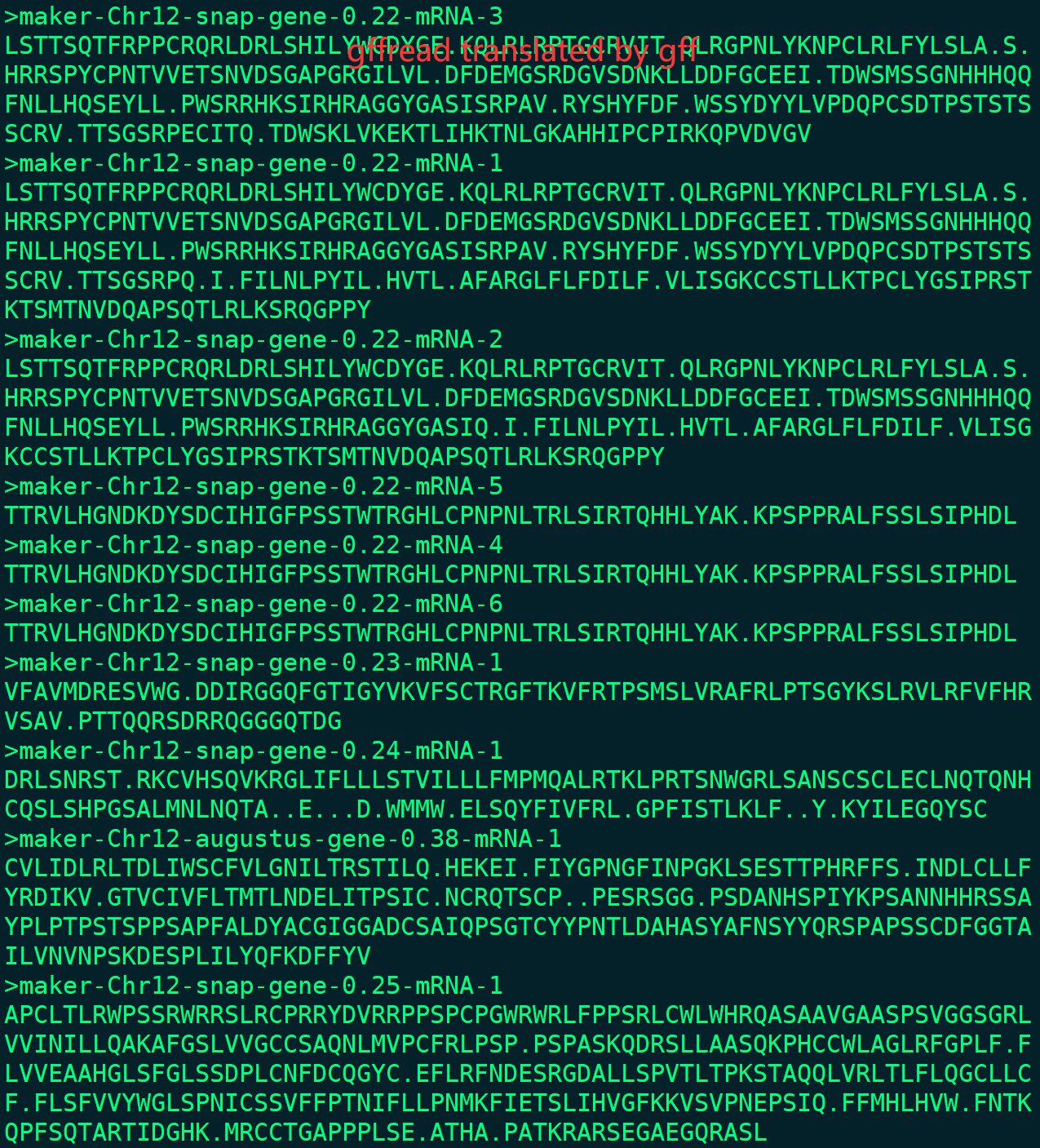
I found a CDS error in my rice project. I ran the maker (3.01.03) and it finished without error in master_datastore_index.log. But when I use gffread to translate the protein from maker gff, I found that almost all of proteins are not start with 'M' and many stop codons in it.
In fact, I checked the protein file (Chr12.maker.proteins.fasta) provided by the maker, it is correct.
I used the same parameter and evidence in another rice, it don't have the problem.
What should I do?
thanks,


Jacques Dainat
Dec 18, 2021, 5:13:33 AM12/18/21
to zc y, maker...@yandell-lab.org
Hi,
Might be related to fragmented CDS where the beginning is missing. The offset (phase) might be different than 0. It is 0 when there is the complete start codon. I don’t know how deals gffread with the offset. You can check within the GFF file if the incriminated genes have this kind of CDS offset start. I know that agat_sp_extract_sequences.pl from AGAT (https://github.com/NBISweden/AGAT) is dealing properly with incomplete CDS and first codon with offset. You might give a try to see if it fix the issue.
Best regards,
Jacques Dainat, Ph.D.
On 29 Nov 2021, at 05:20, zc y <prometh...@gmail.com> wrote:
Dear Maker developers,I found a CDS error in my rice project. I ran the maker (3.01.03) and it finished without error in master_datastore_index.log. But when I use gffread to translate the protein from maker gff, I found that almost all of proteins are not start with 'M' and many stop codons in it.In fact, I checked the protein file (Chr12.maker.proteins.fasta) provided by the maker, it is correct.I used the same parameter and evidence in another rice, it don't have the problem.What should I do?thanks,
<QQ截图20211129121755.png><QQ截图20211129121917.png>_______________________________________________
maker-devel mailing list
maker...@yandell-lab.org
http://yandell-lab.org/mailman/listinfo/maker-devel_yandell-lab.org
Robert King
Jan 12, 2022, 7:41:16 AM1/12/22
to Jacques Dainat, zc y, maker...@yandell-lab.org
If change second from last column to just . then that usually fixes this kind of problem or try script indicated.
Gffread with x parameter and not w right..
From: maker-devel <maker-dev...@yandell-lab.org>
On Behalf Of Jacques Dainat
Sent: 18 December 2021 10:13
To: zc y <prometh...@gmail.com>
Cc: maker...@yandell-lab.org
Subject: Re: [maker-devel] cds error
CAUTION: This email originated from outside of the organisation. Do not click links or open attachments unless you recognise the sender and know the content is safe.
Rothamsted Research is a company limited by guarantee, registered in England at Harpenden, Hertfordshire, AL5 2JQ under the registration number 2393175 and a not for profit charity number 802038.
Reply all
Reply to author
Forward
0 new messages
