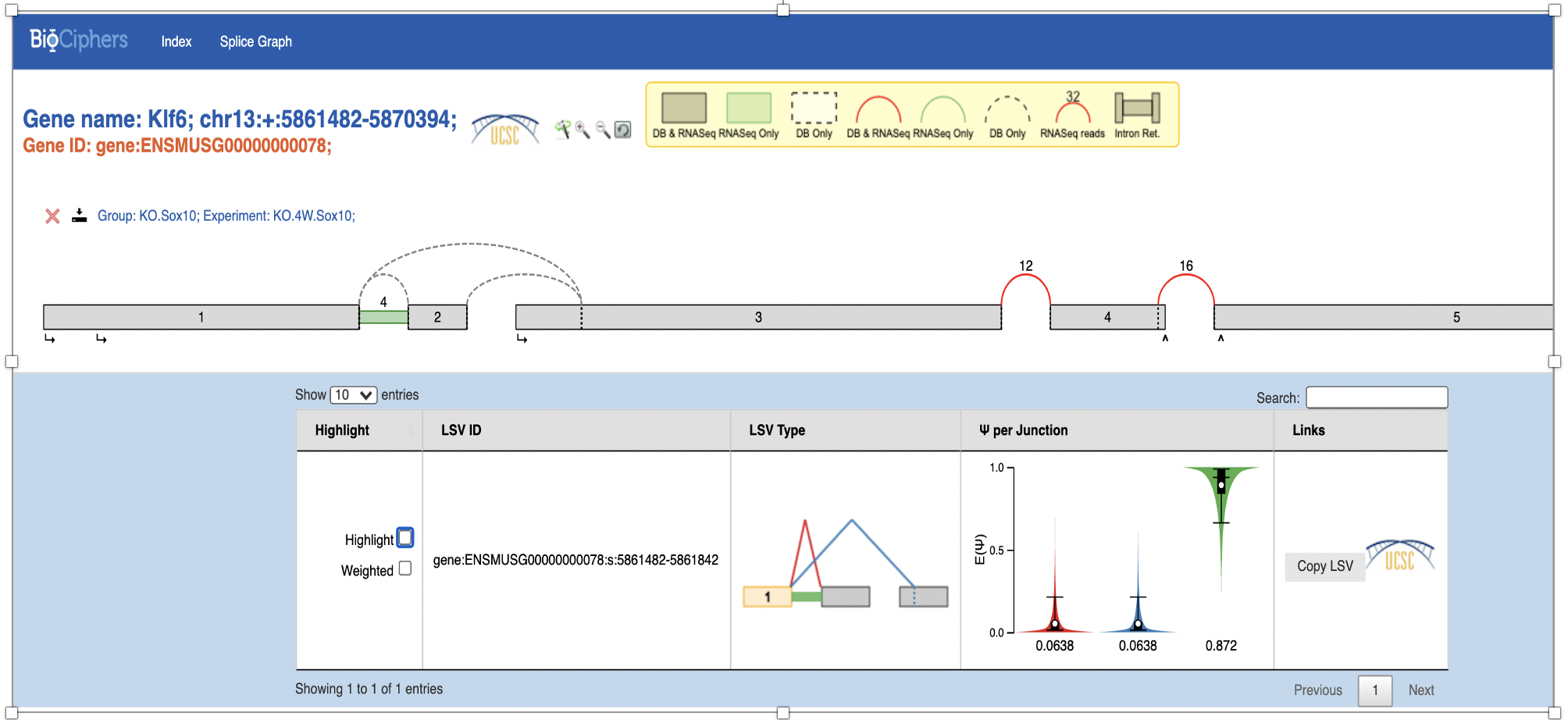
More exons than in annotation file
Justin Malin
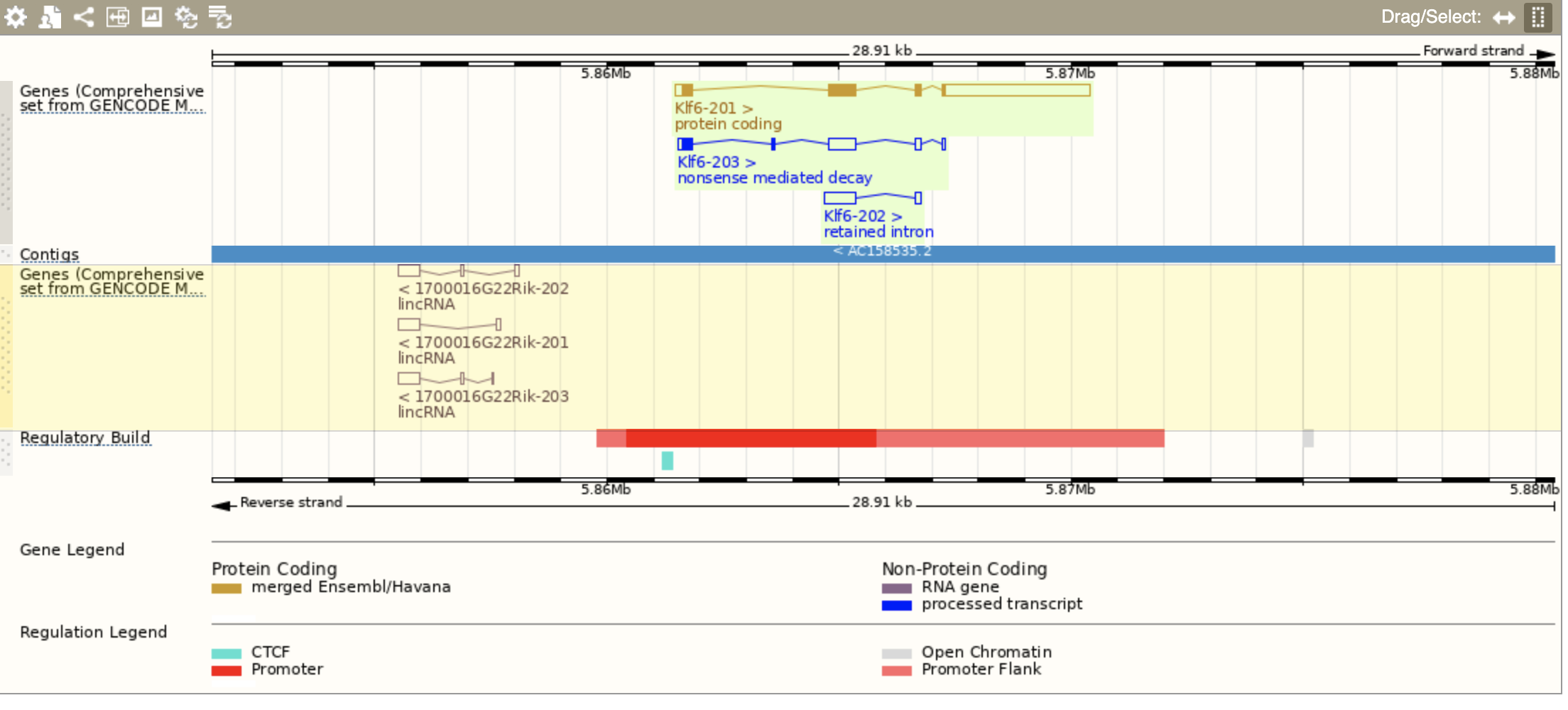
>grep Klf6 Mus_musculus.GRCm38.102.gff3
chr13 ensembl_havana gene 5861482 5870394 . + . ID=gene:ENSMUSG00000000078;Name=Klf6;biotype=protein_coding;description=Kruppel-like factor 6 [Source:MGI Symbol%3BAcc:MGI:1346318];gene_id=ENSMUSG00000000078;havana_gene=OTTMUSG00000066476;havana_version=1;logic_name=ensembl_havana_gene_mus_musculus;version=7
chr13 ensembl_havana mRNA 5861482 5870394 . + . ID=transcript:ENSMUST00000000080;Parent=gene:ENSMUSG00000000078;Name=Klf6-201;biotype=protein_coding;ccdsid=CCDS26229.1;havana_transcript=OTTMUST00000161492;havana_version=1;tag=basic;transcript_id=ENSMUST00000000080;transcript_support_level=1;version=7
chr13 havana mRNA 5861541 5867275 . + . ID=transcript:ENSMUST00000222857;Parent=gene:ENSMUSG00000000078;Name=Klf6-203;biotype=nonsense_mediated_decay;havana_transcript=OTTMUST00000161493;havana_version=1;transcript_id=ENSMUST00000222857;transcript_support_level=5;version=1
chr13 havana lnc_RNA 5864678 5866770 . + . ID=transcript:ENSMUST00000221734;Parent=gene:ENSMUSG00000000078;Name=Klf6-202;biotype=retained_intron;havana_transcript=OTTMUST00000161494;havana_version=1;transcript_id=ENSMUST00000221734;transcript_support_level=2;version=1
Paul Jewell
Justin Malin
Hi Paul,
http://ftp.ensembl.org/pub/release-102/gff3/mus_musculus/Mus_musculus.GRCm38.102.gff3.gz
Thanks,
Justin
Matthew Gazzara
Justin Malin
Hi Matthew,
