DB LSVs not reported in my results
18 views
Skip to first unread message
Bea C
Oct 25, 2021, 4:15:02 AM10/25/21
to majiq_voila
Hello Majiq people!
I am new performing Majiq analysis, and I wondered if anyone could help me to undertand my results. I am analysing 4 RNA-Seq human samples (2 cases and 2 controls). I had no error during the comand line script. But as you can see in the pictures below, the splicegraphs only show few green LSVs per gene.
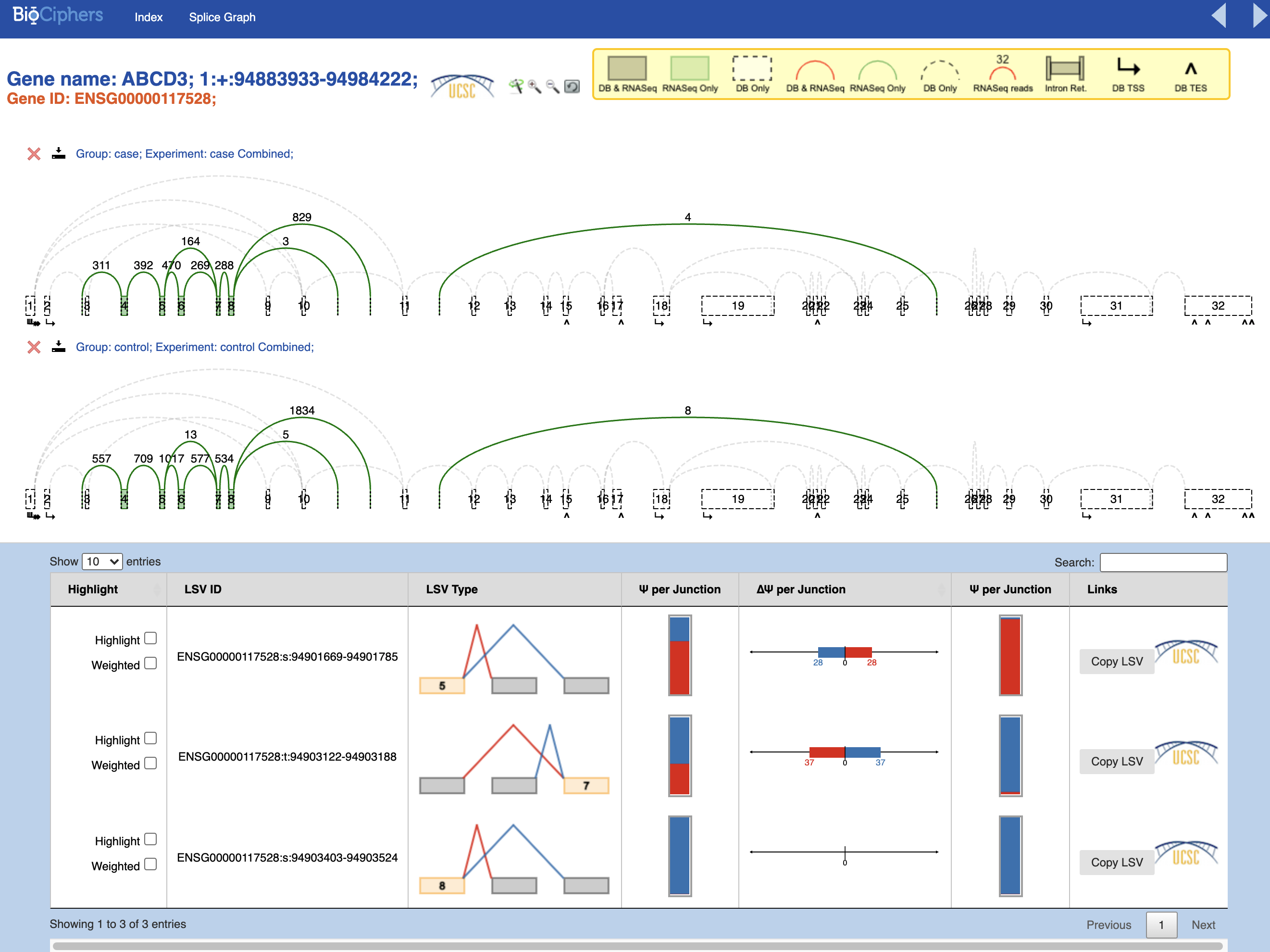
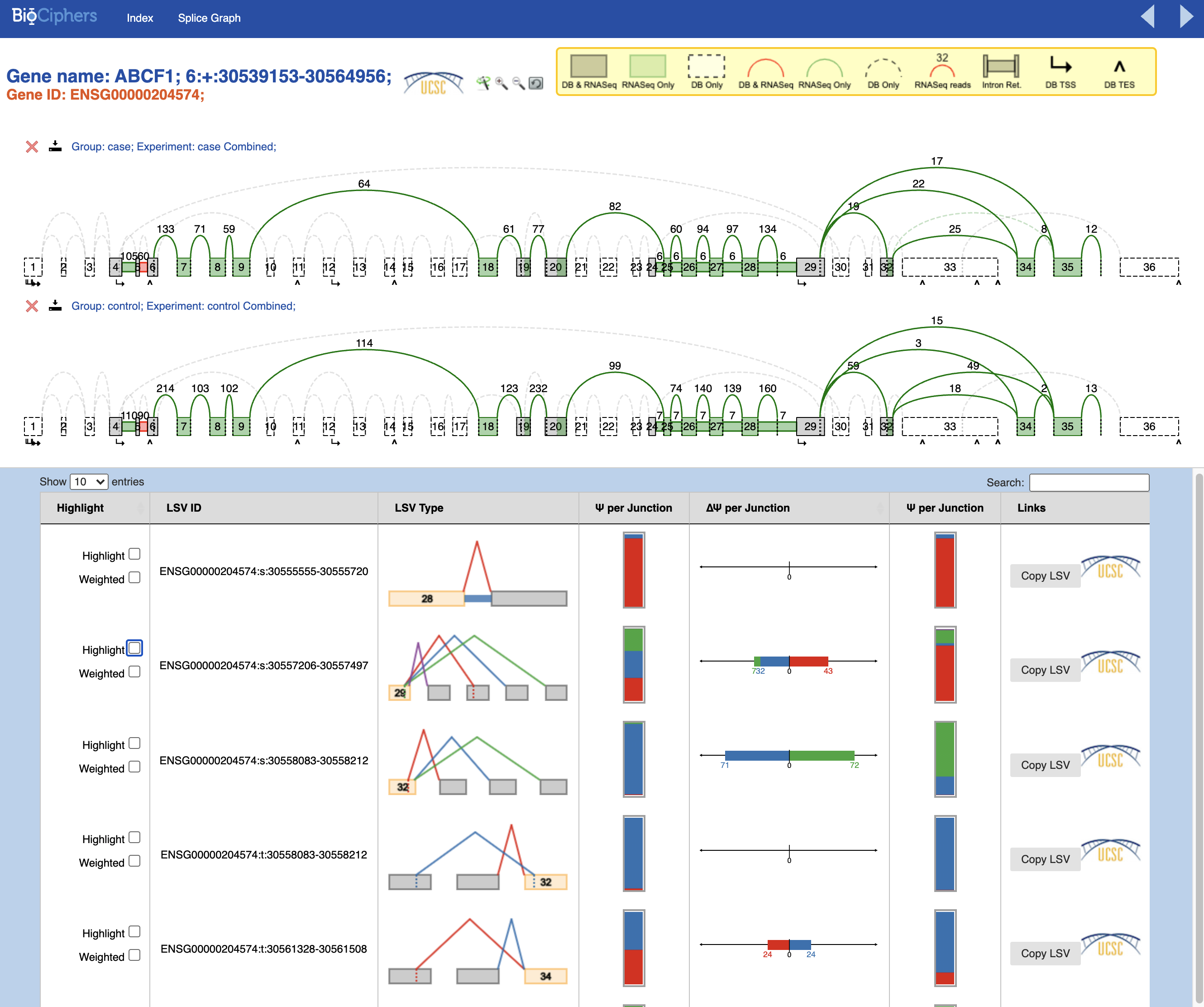
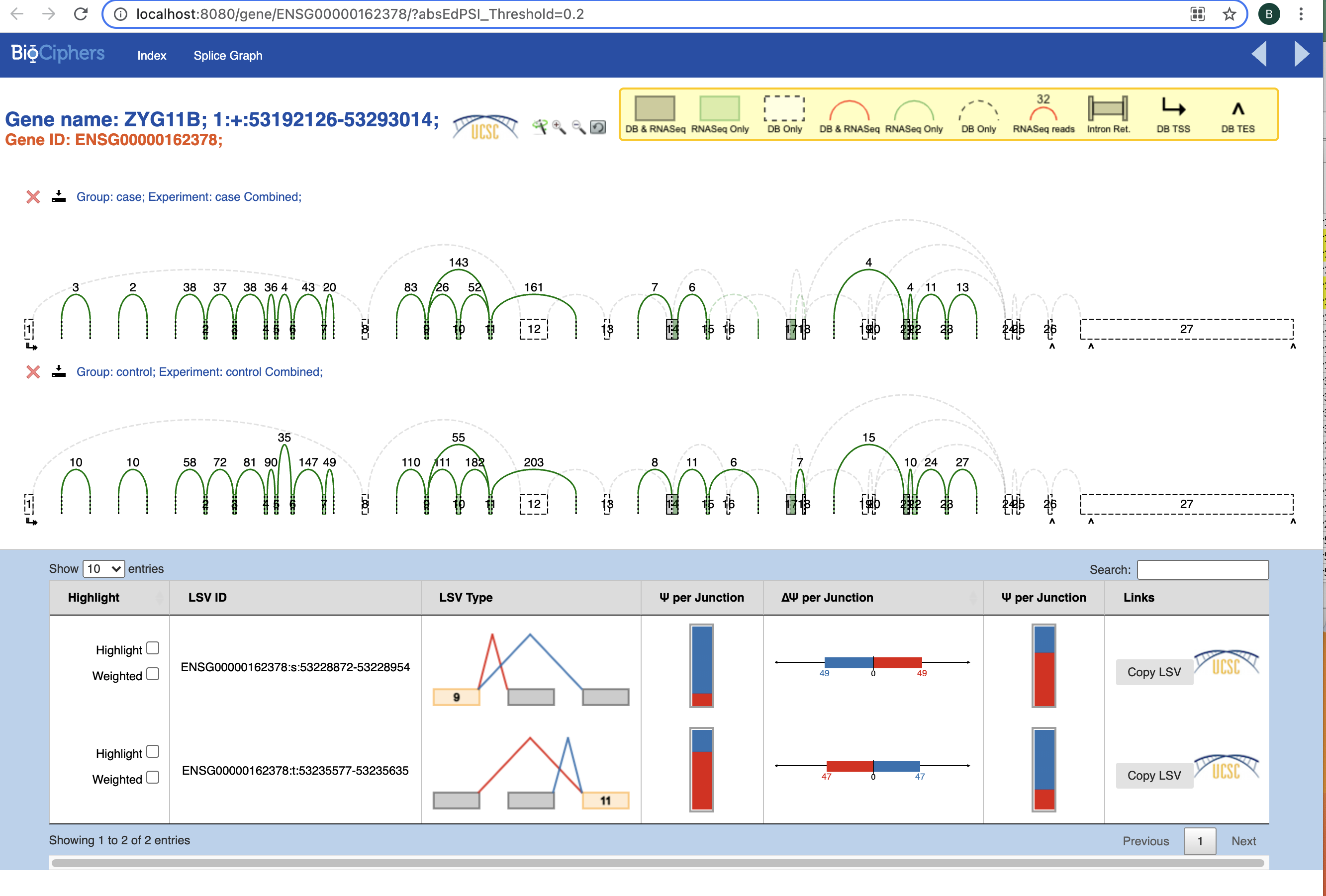
I am new performing Majiq analysis, and I wondered if anyone could help me to undertand my results. I am analysing 4 RNA-Seq human samples (2 cases and 2 controls). I had no error during the comand line script. But as you can see in the pictures below, the splicegraphs only show few green LSVs per gene.
Questions:
1) Why do I have only few LSVs? Is it because the bam files contains reads only for those LSVs?
2) Why the DB LSVs (in red) are not reported? I can see that there are some junctions between two following exons, that should be on DB and reported in red.
Any suggestions? I am using the GFFv3 annotation file recommended by Majiq. Should I use the reference file I used for the alignment?
Any suggestions? I am using the GFFv3 annotation file recommended by Majiq. Should I use the reference file I used for the alignment?
Thank you in advance.
Regards,
Bea



Paul Jewell
Nov 1, 2021, 10:05:43 AM11/1/21
to majiq_voila
Hello Bea,
1) Why are there only a few LSV's: From looking at the reads detected on the provided splicegraphs, the majority of junctions with reads appear constitutive. There will not be LSVs for these.
2) Why are there very few annotated junctions: This depends on the data processed, and the annotation that you have used. The coordinates will need to match. It is not completely impossible to get a result like this though, based on the data used.
Reply all
Reply to author
Forward
0 new messages
