GFF3 error
Juliana Chu
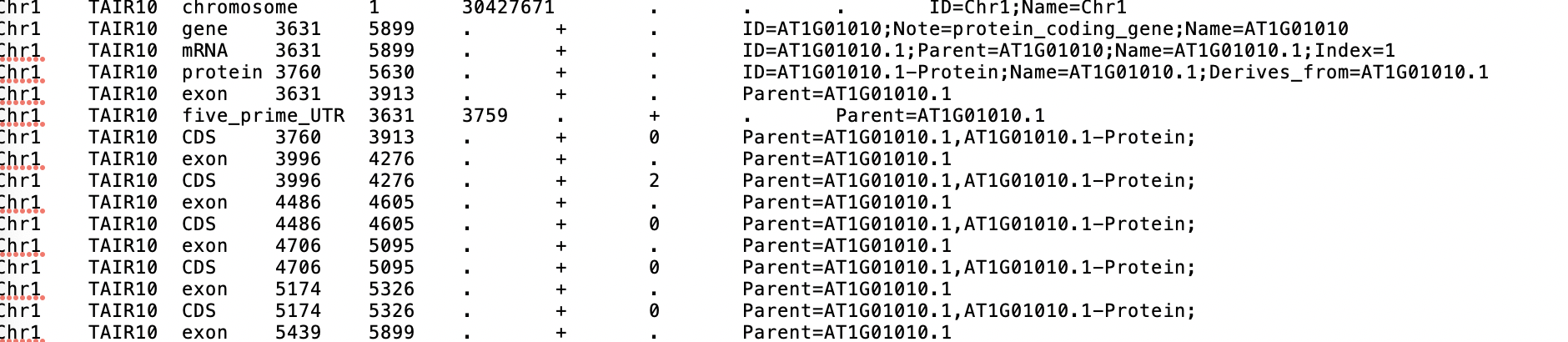
2021-01-06 22:32:18,639 (PID:5889) - INFO - Parsing GFF3
Traceback (most recent call last):
File "/Users/ju.chu/env/bin/majiq", line 11, in <module>
load_entry_point('majiq==2.2', 'console_scripts', 'majiq')()
File "/Users/ju.chu/env/lib/python3.8/site-packages/majiq/run_majiq.py", line 529, in main
args.func(args)
File "majiq/src/build.pyx", line 659, in majiq.src.build.build
File "/Users/ju.chu/env/lib/python3.8/site-packages/majiq/src/basic_pipeline.py", line 13, in pipeline_run
return pipeline.run()
File "majiq/src/build.pyx", line 669, in majiq.src.build.Builder.run
File "majiq/src/build.pyx", line 677, in majiq.src.build.Builder.builder
File "majiq/src/build.pyx", line 580, in majiq.src.build._core_build
File "majiq/src/io.pyx", line 52, in majiq.src.io.read_gff
File "/Users/ju.chu/env/lib/python3.8/site-packages/majiq/src/gff.py", line 74, in parse_gff3
"attributes": __parse_gff_attributes(parts[8]),
File "/Users/ju.chu/env/lib/python3.8/site-packages/majiq/src/gff.py", line 34, in __parse_gff_attributes
key, value = attribute.split("=")
ValueError: not enough values to unpack (expected 2, got 1)
THANK YOU !
Paul Jewell
Juliana Chu
Paul Jewell
Juliana Chu
Hello, Paul!
error: pathspec 'fix-trailing-semicolons-gff3' did not match any file(s) known to git
I don't know what is wrong with the command, since from the first command, it was all done.
Thanks,
-Juliana
Paul Jewell
Joseph Aicher
--
You received this message because you are subscribed to the Google Groups "majiq_voila" group.
To unsubscribe from this group and stop receiving emails from it, send an email to majiq_voila...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/majiq_voila/e120370e-31de-4908-a1b6-001c72d8e314n%40googlegroups.com.
Juliana Chu
remote: Counting objects: 23936, done.
remote: Compressing objects: 100% (5761/5761), done.
remote: Total 23936 (delta 18125), reused 23894 (delta 18097)
Receiving objects: 100% (23936/23936), 21.37 MiB | 536.00 KiB/s, done.
Resolving deltas: 100% (18125/18125), done.
From https://bitbucket.org/biociphers/majiq_academic
* branch HEAD -> FETCH_HEAD
Thanks,
Juliana
Paul Jewell
Hi Juliana,
