Skip to first unread message
Ros
Jun 28, 2020, 7:21:29 AM6/28/20
to lavaan
Hello,

I've posted here before, but am still having a bit of trouble with some errors in my code.
My model is below (N=1282, number of free parameters = 141):
#install.packages("lavaan") #for SEM
library(lavaan)
mymodel <- '
#measurement model
ent =~ sdes_q1 + sdes_q4 + sdes_q6 + sdes_q8
def =~ sdes_q2 + sdes_q3 + sdes_q5 + sdes_q7
idea =~ dssis_q1r + dssis_q2r + dssis_q3r + dssis_q4r
rIP =~ sresilience_q1 + sresilience_q2 + sresilience_q5 + sresilience_q9 + sresilience_q11 + sresilience_q13 + sresilience_q19 + sresilience_q20 + sresilience_q25
rES =~ sresilience_q4 +sresilience_q8 + sresilience_q10 + sresilience_q12 + sresilience_q14 + sresilience_q18 + sresilience_q23 + sresilience_q24
rEP =~ sresilience_q3 + sresilience_q6 + sresilience_q7 + sresilience_q11 + sresilience_q13 + sresilience_q15 + sresilience_q16 + sresilience_q17 + sresilience_q21 + sresilience_q22
bur =~ inq_q1 + inq_q2 + inq_q3 + inq_q4 + inq_q5
belonging =~ inq_q6r + inq_q7r + inq_q8 + inq_q9 + inq_q10r
dep =~ cessds_q1 + cessds_q2 + cessds_q3 + cessds_q4 + cessds_q5r + cessds_q6 + cessds_q7 + cessds_q8r + cessds_q9 + cessds_q10
#structural model
def ~ dep
ent ~ def + dep
idea ~ ent + bur + bel + rEP + rES + rIP + dep'
# model identification and estimation
modelfit <- sem(mymodel, data = mydatat1, estimator = "MLM")
summary(modelfit_ideation, fit.measures = T, standardized = T, rsquare = T)
I get two errors in my output:
1. lavaan WARNING: some estimated lv variances are negative
I should note that the model above is a direct replication study of a published SEM in MPlus (but using a different sample). My current understanding is that this error may be a result of a misspecified model, but I can't say for sure.
and
2. When I try to print the R square values for my latent variables, I get NA for one of them.
These errors don't happen when I include a different outcome variable that is binary ('ordered') and specify my model estimation as DWLS.
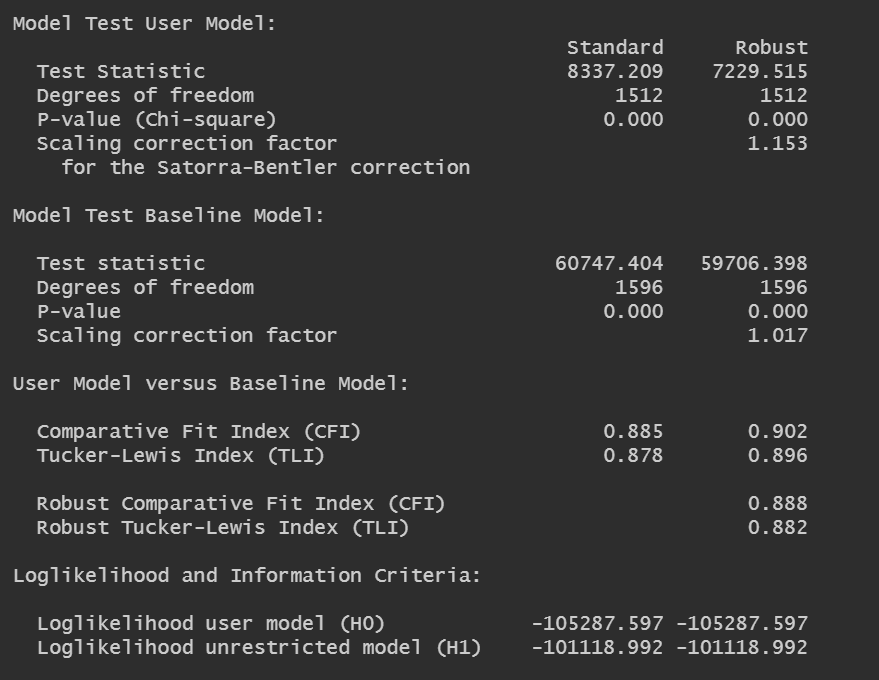
In the output, I also seem to get two columns worth of values (e.g. fo CFI and TLI), and I can't seem to find guidance as to which one to take note of:
I'm quite new to this, and I'd really appreciate some guidance on these errors, and what I could be doing wrong.
Many thanks!
Patrick (Malone Quantitative)
Jun 28, 2020, 9:14:19 AM6/28/20
to lav...@googlegroups.com
Ros,
I might be able to help a little.
For point 1, check the residual variances section. I'm betting you'll find that "ent" is the latent with the negative residual. That would explain point 2, because the R^2 is undefined (or could be reported as >1, also unallowable) if more variance in ent is accounted for than exists.
For point 3, you requested MLM in your sem() call, which is the Satorra-Bentler adjusted ("robust") chi-square. Since you asked for the robust statistic, I would think that's the column you want to look at for chi-square and statistics based on it e..g., CFI), even though lavaan returns the unadjusted/standard as well.
Pat
--
You received this message because you are subscribed to the Google Groups "lavaan" group.
To unsubscribe from this group and stop receiving emails from it, send an email to lavaan+un...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/lavaan/64d64c64-7b17-4938-bb76-47494fb91268o%40googlegroups.com.
--
Patrick S. Malone, Ph.D., Malone Quantitative
NEW Service Models: http://malonequantitative.com
He/Him/His
NEW Service Models: http://malonequantitative.com
He/Him/His
Rosie Pendrous
Jun 28, 2020, 12:18:02 PM6/28/20
to lav...@googlegroups.com
Hi Pat, that's really helpful thank you. You're right in that the residual variance value only for ent is negative. Do you perhaps also know how I could resolve this issue?
To view this discussion on the web visit https://groups.google.com/d/msgid/lavaan/CAJc%3DyOESAjbmRoWfJhdWqBpOv2DxOAWQJSHa_p%3Drmet_AK19_A%40mail.gmail.com.
Patrick (Malone Quantitative)
Jun 28, 2020, 1:45:09 PM6/28/20
to lav...@googlegroups.com
This is called a "Heywood case," which should give you a start on web-searches about causes and remedies. There's a lot out there, particularly on stackexchange.
To view this discussion on the web visit https://groups.google.com/d/msgid/lavaan/CANc%2B4-CiKFRTCvrUxTYbWjp_H6-CKj2%2ByDmSzPUxGggHmm9q3w%40mail.gmail.com.
Reply all
Reply to author
Forward
0 new messages
