Not sure if it is a bug of the colon operator or if it is a known limitation. It should work if you compute the product term yourself. As far as I know, the colon operator just forms the product terms internally. What it does should be equivalent to forming the product term ourselves.
I can reproduce the error using a toy dataset:
library(lavaan)
#> This is lavaan 0.6-9
#> lavaan is FREE software! Please report any bugs.
set.seed(4314)
n <- 100
B <- rnorm(n)
IV <- rnorm(n)
A <- rnorm(n)
Outcome <- sample(c("a", "b"), n, replace = TRUE)
df <- data.frame(A, B, IV, Outcome)
df$IVB <- df$IV * df$B
mod.med <- '
#Regression
A~ a1*IV + a2*B + a3*IV:B
Outcome~ c1*IV+ b*A
'
my.fit <- sem(mod.med, data=df, missing = 'listwise', bootstrap = 10, se = "bootstrap", ordered ="Outcome", estimator = "DWLS")
#> Warning in lav_options_set(opt): lavaan WARNING: information will be set to
#> "expected" for estimator = "DWLS"
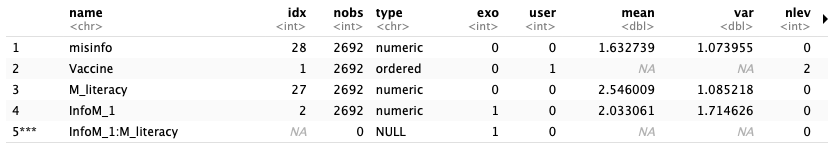
#> name idx nobs type exo user mean var nlev lnam
#> 1 A 1 100 numeric 0 0 -0.02342639 0.9369112 0
#> 2 Outcome 4 100 ordered 0 1 NA NA 2 a|b
#> 3 IV 3 100 numeric 1 0 -0.13189269 1.0606386 0
#> 4 B 2 100 numeric 1 0 0.05082713 0.9287101 0
#> 5*** IV:B NA 0 NULL 1 0 NA NA 0
#> Error in lav_data_full(data = data, group = group, cluster = cluster, : lavaan ERROR: some variables have no values (only missings) or no variance
(P.S.: I am not sure why IV[M_literacy] is endogenous in your output but endogenous in the example above.)
The following version use IVB, computed before calling sem():
library(lavaan)
#> This is lavaan 0.6-9
#> lavaan is FREE software! Please report any bugs.
set.seed(4314)
n <- 100
B <- rnorm(n)
IV <- rnorm(n)
A <- rnorm(n)
Outcome <- sample(c("a", "b"), n, replace = TRUE)
df <- data.frame(A, B, IV, Outcome)
df$IVB <- df$IV * df$B
mod.med <- '
#Regression
A~ a1*IV + a2*B + a3*IVB
Outcome~ c1*IV+ b*A
'
my.fit <- sem(mod.med, data=df, missing = 'listwise', bootstrap = 10, se = "bootstrap", ordered ="Outcome", estimator = "DWLS")
#> Warning in lav_options_set(opt): lavaan WARNING: information will be set to
#> "expected" for estimator = "DWLS"
summary(my.fit)
#> lavaan 0.6-9 ended normally after 10 iterations
#>
#> Estimator DWLS
#> Optimization method NLMINB
#> Number of model parameters 8
#>
#> Number of observations 100
#>
#> Model Test User Model:
#>
#> Test statistic 1.902
#> Degrees of freedom 2
#> P-value (Chi-square) 0.386
#>
#> Parameter Estimates:
#>
#> Standard errors Bootstrap
#> Number of requested bootstrap draws 10
#> Number of successful bootstrap draws 10
#>
#> Regressions:
#> Estimate Std.Err z-value P(>|z|)
#> A ~
#> IV (a1) -0.141 0.059 -2.379 0.017
#> B (a2) -0.017 0.102 -0.166 0.868
#> IVB (a3) -0.135 0.080 -1.680 0.093
#> Outcome ~
#> IV (c1) -0.096 0.179 -0.536 0.592
#> A (b) -0.136 0.108 -1.265 0.206
#>
#> Intercepts:
#> Estimate Std.Err z-value P(>|z|)
#> .A -0.016 0.087 -0.183 0.855
#> .Outcome 0.000
#>
#> Thresholds:
#> Estimate Std.Err z-value P(>|z|)
#> Outcome|t1 -0.058 0.067 -0.862 0.388
#>
#> Variances:
#> Estimate Std.Err z-value P(>|z|)
#> .A 0.894 0.099 8.987 0.000
#> .Outcome 0.983
#>
#> Scales y*:
#> Estimate Std.Err z-value P(>|z|)
#> Outcome 1.000
Created on 2022-01-28 by the reprex package (v2.0.1)
By the way, was DWLS specified explicitly because you want to get bootstrapping CIs? As shown above, robust test statistics, normally reported as in the example at
https://lavaan.ugent.be/tutorial/cat.html if ordered is set, will not be computed if we specify estimator = "DWLS" directly.
If you really want to do what the example does but also need bootstrapping CIs, you can add test = "scaled.shifted":
my.fit <- sem(mod.med, data=df, missing = 'listwise', bootstrap = 10, se = "bootstrap", ordered ="Outcome", estimator = "DWLS", test = "scaled.shifted")
#> Warning in lav_options_set(opt): lavaan WARNING: information will be set to
#> "expected" for estimator = "DWLS"
summary(my.fit)
#> lavaan 0.6-9 ended normally after 10 iterations
#>
#> Estimator DWLS
#> Optimization method NLMINB
#> Number of model parameters 8
#>
#> Number of observations 100
#>
#> Model Test User Model:
#> Standard Robust
#> Test Statistic 1.902 1.934
#> Degrees of freedom 2 2
#> P-value (Chi-square) 0.386 0.380
#> Scaling correction factor 0.986
#> Shift parameter 0.004
#> simple second-order correction
#>
#> Parameter Estimates:
#>
#> Standard errors Bootstrap
#> Number of requested bootstrap draws 10
#> Number of successful bootstrap draws 10
Created on 2022-01-28 by the reprex package (v2.0.1)
However, I would not do this because there may be other default options that I may forget to set for analyzing endogenous ordered categorical variable (e.g, the warning displayed above).
A better approach is to omit estimator, se, and test, as in the example in
https://lavaan.ugent.be/tutorial/cat.html, and let lavaan set up the options for us. Request bootstrap CIs in a separate call to sem(), as you did in the original code, if you need them.
My two cents.
-- Shu Fai