Converting multi-animal, multi-part DeepLabCut CSV to JAABA-compatible track files
Kevin Chen
Mayank Kabra
--
You received this message because you are subscribed to the Google Groups "JAABA" group.
To unsubscribe from this group and stop receiving emails from it, send an email to jaaba+un...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/jaaba/b8073d6f-861f-4920-9aec-c710a9e6c962n%40googlegroups.com.
Mayank Kabra
Hi Mayank,Could you elaborate more on why we need to fix those fields? The output of DeepLabCut returns the locations of the body parts of both rats, but we don't have the position of every rat for every frame. As such, we included the frames within the first and last frames that we have a position. As a result, the values for firstframe, endframe, and nframes differ between the two rats. Is that the issue you were referring to? Because of these gaps in the tracking, the dt is also not always consistent.Thank you for your help and expertise,Kevin Chen & Etienne RichartOn Wed, Sep 22, 2021 at 2:39 AM Mayank Kabra <mayank...@gmail.com> wrote:Hi Kevin,Ah, sorry didn't realize that. Yes, the p.mat file's format is wrong. The details of trx files format can be found here: http://jaaba.sourceforge.net/DataFormatting.html#TracksFileStructure . You will have to update the conversion code to comply with the trx file format. Specifically, fields like firstframe, lastframe, and off.MayankOn Tue, Sep 21, 2021 at 9:55 PM Kevin Chen <che...@carleton.edu> wrote:Hi,First of all, thank you so much for taking the time to help us out, we really appreciate it!It seems that there may have been a misunderstanding; the registered_trx.mat file is the template that we used to create the new tracks file: p.mat. We changed the "trx" struct within registered_trx.mat to a struct that contains our data( based off of JAABA's documentation for preparing data), but didn't change the "timestamps" field.We are unsure on what "timestamps" does, could you let us know what we should put for that?To be clear, the tracks file that we used is called "p.mat", not "registered_trx.mat".Thanks again for your help,Kevin ChenCarleton College '23Computer Science & Statistics
Kevin Chen
Hello Mayank,
Thanks for your earlier advice. Once we implemented the changes you suggested we got past the original issue but are now running into a new one. We have updated the drive we previously shared with the new conversion code (“trackConverter2.py”), a new trx.mat file ( “p.mat”) and a .mp4 version of our movie ( “movie.mp4”) as well as a folder ( testBehavior1) which is our experiment directory that we load into JAABA. The link to the files if you need it: https://drive.google.com/drive/folders/1Z1rHoflMsMsN9Lk8C_TN2sSugRYFP9M_?usp=sharing
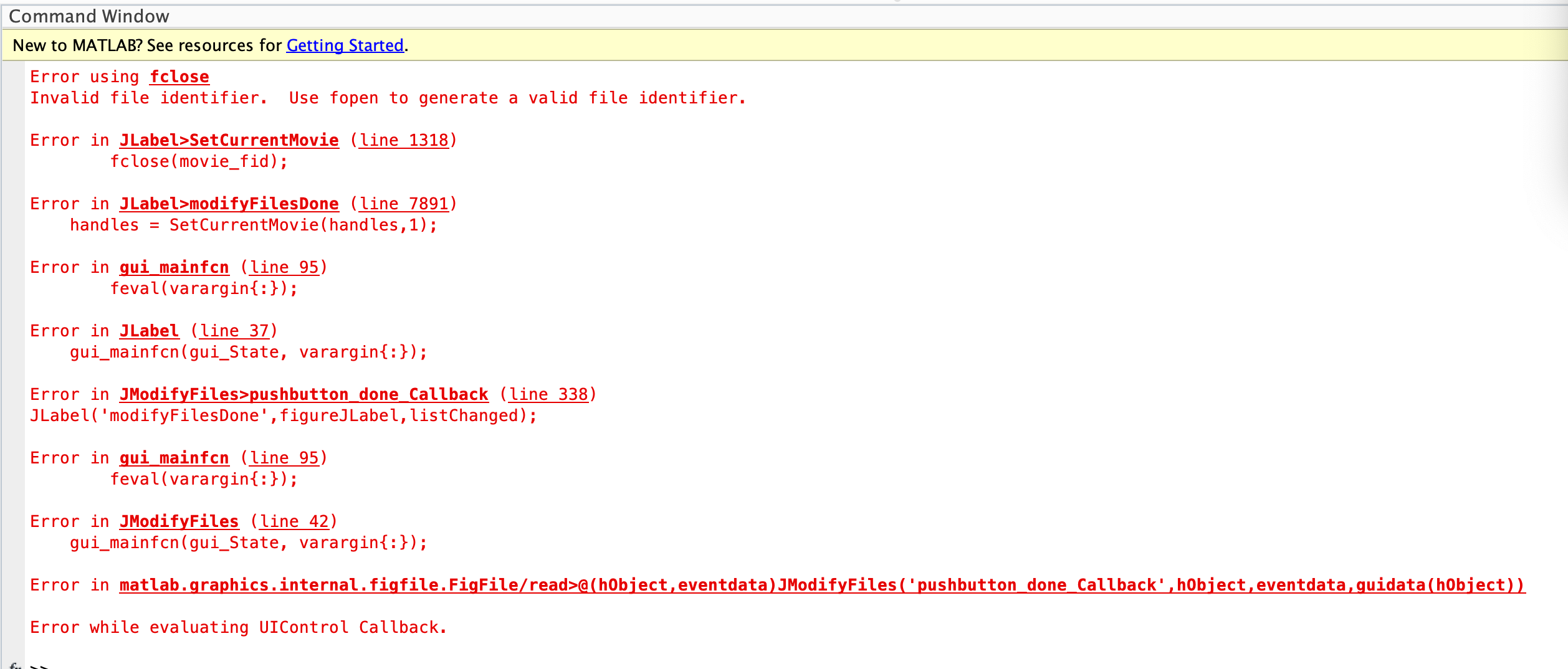
When we now try to create a new project we can load the movie, but before we get past the black screen, the software throws an error, which we have included below.
Do you have any suggestions for how we can resolve this error? We believe it could be a problem with how the trx.mat data is formatted. We’ve checked the dimensions of the fields in the output file (“p.mat”) thoroughly and don’t see an issue. We also speculate that perhaps our extensive use of NaN values might be the cause of the error; we’ve had some difficulty in converting our DeepLabCut tracks into some of the features in the trx.mat file because the documentation is vague. For instance, we were unsure on what values to put in for “theta”, “x_mm”, “theta_mm”.
Also, we believe the issue might also originate from the feature config file(featureConfig_logan_rat.xml). We were given that xml file from the previous person who worked on this project, but we don’t know what any of it means. Is there some documentation on how the xml file should be formatted?
Thanks again for your help.
Sincerely,
Kevin Chen & Etienne Richart

Mayank Kabra
To view this discussion on the web visit https://groups.google.com/d/msgid/jaaba/77bb2c9b-2f90-4360-a038-db2a58712323n%40googlegroups.com.
