How to generate new segmentations from an existing segmentation
Daniel
Cook, Philip
I'm not sure how the second and third segmentations differ. You can dilate a segmentation with c3d. Assuming you have a binary ROI with label 1 for hematoma, 0 for background
c3d hematoma.nii.gz -as original -dilate 1 2x2x2mm -push original -scale -1 -add -o newROI.nii.gz
This will give you a 2mm border around the original label.
--
You received this message because you are subscribed to the Google Groups "itksnap-users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to
itksnap-user...@googlegroups.com.
To view this discussion on the web visit
https://groups.google.com/d/msgid/itksnap-users/815de1c9-f355-46a2-bbea-8362c8493046n%40googlegroups.com.
Daniel
Cook, Philip
On Nov 3, 2020, at 7:10 PM, Daniel <yan...@gmail.com> wrote:
Thank you for the prompt reply, it helps a lot!
To view this discussion on the web visit https://groups.google.com/d/msgid/itksnap-users/bed516f5-59e4-4044-987a-cfd025811239n%40googlegroups.com.
Daniel
Thank you. I will try both commands!
Daniel
Cook, Philip
On Nov 4, 2020, at 11:39 PM, Daniel <yan...@gmail.com> wrote:
I have tried both commands, and it seems they are not working as intended.
For example, when using the "dilate"command, the generated segmentation extends from the original segmentation more far than the value I set.(If I set the value to be 5 mm, for example, the distance between the border of the generate segmentation and the border of the original segmentation is visually much greater than 5 mm.)
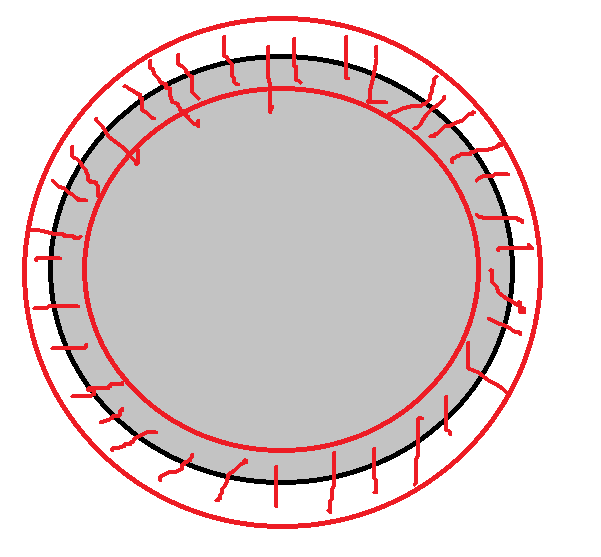
What I want is drawing the border of a lesion (the original segmentation, which i already have), and then extends the border inward for 5 mm, and outward for 5 mm.And finally I can get an area on the border of the lesion, the center-line of which is the actual border, and the width of the area is 10 mm.
Following is an illustration of what I want.I have the black circle already (the original segmentation), and I want to get the red circular area (the actual shape of the area depends on the original segmentation) that centers on the black circle, the width of which is 10 mm.
To view this discussion on the web visit https://groups.google.com/d/msgid/itksnap-users/40e55dce-2b5a-474d-98bf-4be4e0aeeb8en%40googlegroups.com.
<无标题.png>
Dorian P.
To view this discussion on the web visit https://groups.google.com/d/msgid/itksnap-users/8119F784-ED24-4D0B-9F69-E5C7C88D4CF7%40pennmedicine.upenn.edu.
Daniel
Daniel
Cook, Philip
I don't need the DICOM, just the NIFTI segmentation, which should be quite small if compressed as .nii.gz. I can send you a link where you can upload it.
To view this discussion on the web visit https://groups.google.com/d/msgid/itksnap-users/70e7908e-8f97-48f9-83b8-98d688c6e733n%40googlegroups.com.
Daniel
Daniel
Daniel
Olena Tankyevych
To view this discussion on the web visit https://groups.google.com/d/msgid/itksnap-users/64b16f07-fd62-453f-80b8-e104c40c59a4n%40googlegroups.com.
Cook, Philip
On Nov 19, 2020, at 2:58 AM, Daniel <yan...@gmail.com> wrote:
Hi Philip,
To view this discussion on the web visit https://groups.google.com/d/msgid/itksnap-users/64b16f07-fd62-453f-80b8-e104c40c59a4n%40googlegroups.com.
