Strange Scores on Reads in a Predicted Intron Site
16 views
Skip to first unread message
Mason Lee
Jun 29, 2022, 10:59:35 AM6/29/22
to Integrated Genome Browser Help Desk
Hello,
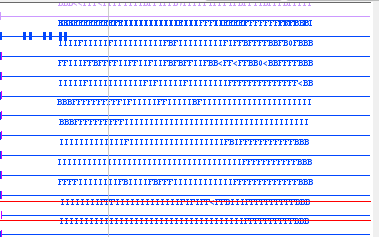
I am working with a HISAT2 alignment.
Is anyone able to interpret these results? It should be noted that this is an intron region of considerable predicted length (~7000 bp). These long reads are given with scores corresponding to these letters. I assume these are not true?

Nowlan Freese
Jul 7, 2022, 11:25:00 AM7/7/22
to Integrated Genome Browser Help Desk
Hi Mason,
It looks like the Label Field is set to "scores" in IGB. Assuming that is the case, IGB will show the score of the reads up to a character limit centered on the read itself.
If you are interested in comparing Phred scores between reads then right-clicking on the track label and selecting Shade base by quality may work better. If you want the score for any particular read you can click to select the read and then select the Selection Info tab which should contain the scores field.
Let me know if this helps.
Best,
Nowlan
Reply all
Reply to author
Forward
0 new messages
