IDR problem with ChIPmentation data
139 views
Skip to first unread message
Frédéric Crémazy
Oct 29, 2020, 10:06:32 AM10/29/20
to idr-discuss
Hello,
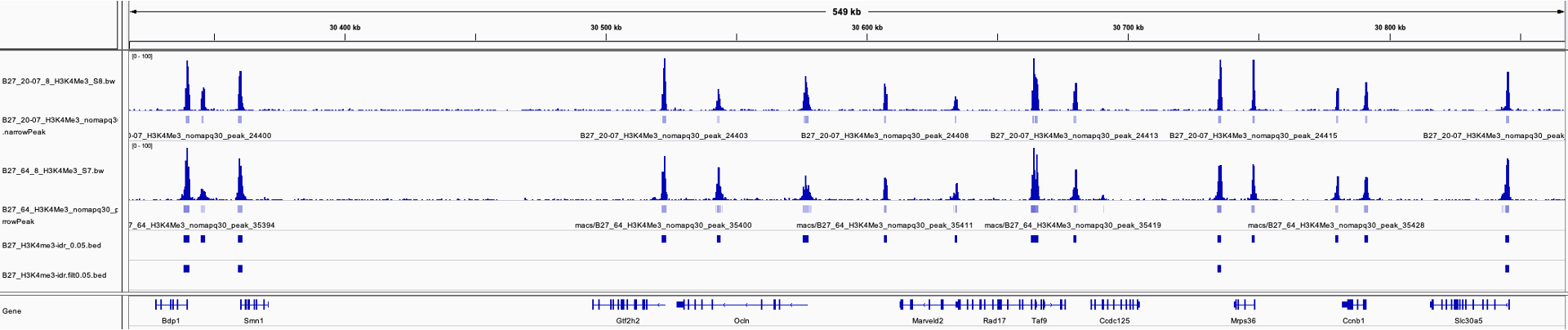
I am trying to use IDR on Histone mods ChIPmentation data (i.e. H3K4me3 and H3K27ac) to extract relevant peaks from replicate samples.
Running IDR on 2 narrow peak files by doing:
idr --samples B27_20-07_H3K4Me3_peaks.narrowPeak B27_64_H3K4Me3_peaks.narrowPeak --input-file-type narrowPeak --rank p.value --output-file B27_H3K4me1-idr --plot --log-output-file B27_H3K4me1-idr.log
gives me these results:
IDR cutoff 0.05
•Initial parameter values: [0.10 1.00 0.20 0.50]
•Final parameter values: [1.36 0.61 0.52 0.24]
•Number of reported peaks - 28274/28274 (100.0%)
•Number of peaks passing IDR cutoff of 0.05 - 1553/28274 (5.5%)
I think 5.5% is very low compared to how reproducible look my 2 profiles and when I compare on IGV the 2 original files with a file containing only the peaks that passed the 0.05 cutoff I really have the impression that something is wrong (see attached image, IDR results on the last line).
Can you help to understand what is happening here and what I am doing wrong?
Thank you very much for your help,
Frédéric

Anshul Kundaje
Oct 29, 2020, 10:19:09 AM10/29/20
to idr-d...@googlegroups.com
We don't recommend using IDR with histone marks and MACS2 as the peak caller. Instead just use peaks that overlap between replicates or pseudo-replicates. That works well.
IDR works well with punctate events like TF binding or chromatin accessibility and peak callers that have smooth peak ranking measures.
Anshul
--
You received this message because you are subscribed to the Google Groups "idr-discuss" group.
To unsubscribe from this group and stop receiving emails from it, send an email to idr-discuss...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/idr-discuss/d43efa02-ceab-4279-8d4e-8951195dfd08n%40googlegroups.com.
Reply all
Reply to author
Forward
0 new messages
