Question about Active Brownian Particles
koji iwase
I am Koji Iwase, a master course graduate student (Nagoya Institute of Technology) in Japan.
I am planning to simulate Active Brownian Particles (ABPs) in a two-dimensional system. ABPs are represented as follows.
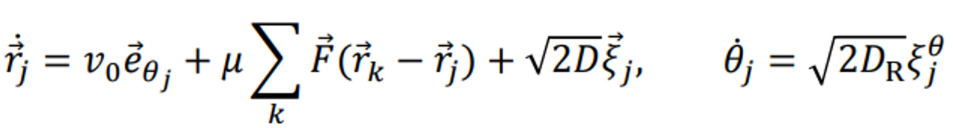
Does the code shown above accurately represent the ABP equation?
(The above code was implemented with reference to the example_active.py in this post (https://groups.google.com/g/hoomd-users/c/mqpbfUQd-DY/m/sRBF0zZeAQAJ))
What I particularly don't understand is:
```
rotational_diffusion_updater = active.create_diffusion_updater(
trigger=hoomd.trigger.Periodic(1), rotational_diffusion=DR
)
```
brownian.gamma_r.default =[gamma_r,gamma_r,0]
```
Here, the variables related to rotational diffusion(gamma_r、DR) are being set twice.
In the above code, it seems that the updating of the rotation angle θ is being done twice.
Also, according to the translational diffusion section of Brownian's documentation (https://hoomd-blue.readthedocs.io/en/v3.11.0/module-md-methods.html#hoomd.md.methods.Brownian)...
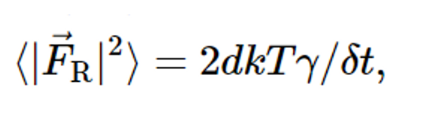
It says that the third term of ABPs show below cannot be matched, considering that my system is two-dimensional, so d (dimensionality of the system) = 2.
(D=kbT/gamma)
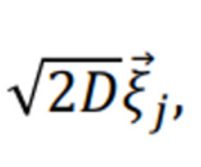
I apologize for my lack of knowledge and for asking a clumsy question. Thank you in advance for your response.
koji iwase
Could you give me some reaction?
Joshua Anderson
On May 13, 2023, at 10:35 AM, koji iwase <koji.iwa...@gmail.com> wrote:
Could you give me some reaction?
2023年5月9日火曜日 14:49:35 UTC+9 koji iwase:
Dear all,
I am Koji Iwase, a master course graduate student (Nagoya Institute of Technology) in Japan.
I am planning to simulate Active Brownian Particles (ABPs) in a two-dimensional system. ABPs are represented as follows.
<タイトルなし.png>
Does the code shown above accurately represent the ABP equation?
(The above code was implemented with reference to the example_active.py in this post (https://groups.google.com/g/hoomd-users/c/mqpbfUQd-DY/m/sRBF0zZeAQAJ))
What I particularly don't understand is:
```
rotational_diffusion_updater = active.create_diffusion_updater( trigger=hoomd.trigger.Periodic(1), rotational_diffusion=DR )
```
brownian.gamma_r.default =[gamma_r,gamma_r,0]
```
Here, the variables related to rotational diffusion(gamma_r、DR) are being set twice.
In the above code, it seems that the updating of the rotation angle θ is being done twice.
Also, according to the translational diffusion section of Brownian's documentation (https://hoomd-blue.readthedocs.io/en/v3.11.0/module-md-methods.html#hoomd.md.methods.Brownian)...
<タイトルなし2.png>
It says that the third term of ABPs show below cannot be matched, considering that my system is two-dimensional, so d (dimensionality of the system) = 2.
(D=kbT/gamma)
<タイトルなし3.png>
I apologize for my lack of knowledge and for asking a clumsy question. Thank you in advance for your response.
--
You received this message because you are subscribed to the Google Groups "hoomd-users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to hoomd-users...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/hoomd-users/d48f09c9-b22e-428a-a8cf-84d0375b74cdn%40googlegroups.com.
<タイトルなし2.png><タイトルなし.png><タイトルなし3.png>
Brandon Butler
Koji,
The Active force does not set the "anisotropic" flag, so the
particles will be treated as point particles. As such they will
have 0 torque and angular momentum with the Brownian integrator
method. You could set the integrator flag integrate_rotational_dof
to True and give your particles a moment of inertia to use
the Brownian method's rotational diffusion if desired, but then
you do not need to use the ActiveRotationalDiffusion updater. The
active rotational diffusion will give you give diffusion of the
direction of force in active particles without needing the to do
the aforementioned actions.
Best,
Brandon
--
You received this message because you are subscribed to the Google Groups "hoomd-users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to hoomd-users...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/hoomd-users/d48f09c9-b22e-428a-a8cf-84d0375b74cdn%40googlegroups.com.
Brandon Butler
MolSSI Fellow
PhD Candidate, Chemical Engineering and Scientific Computing | Glotzer Lab, University of Michigan
Email: butl...@umich.edu
