Creation of Fixed Particle walls
Russell Holt
Hi all
I am new to HOOMD-Blue however have some experience in other MD packages (LIGGGHTS/LAMMPS). I have recently installed the Fast Stokesian Dynamics (FSD) package ‘PSEv3’ and am interested in applying this package to a pressure driven channel flow.
I have run the included test simulation which comes with the package which uses shearing walls to imply a fluid flow, however what I would like to do is a little different and I’m not sure if this is something HOOMD can do.
I would like to have fixed particles which make up the walls of the channel, and then have a separate flowing group. I believe I should be able to use the ‘rigid’ constraint on the wall particles and then apply the PSEv3 to resolve the hydrodynamic interactions, (with Stokesian Dynamics, the fixed wall particles are required to be included in a mobility matrix which describes the flow of the particles moving through the channel, thus the wall needs to be particles). My problem at the moment is that I cannot see how to make these separate groups for the wall particles. Can the init.read_getar command be used to read in a set of particles, fix these and then create a lattice of particles for the flowing group?
A simple 2D
example of this system being run in LAMMPS can be seen attached, where the white
particles on the top and bottom are effectively ‘frozen’, still allowing
interactions to be calculated, however themselves not moving. This will create the desired parabolic poiseulle velocity profile when a pressure gradient is applied across all particles. 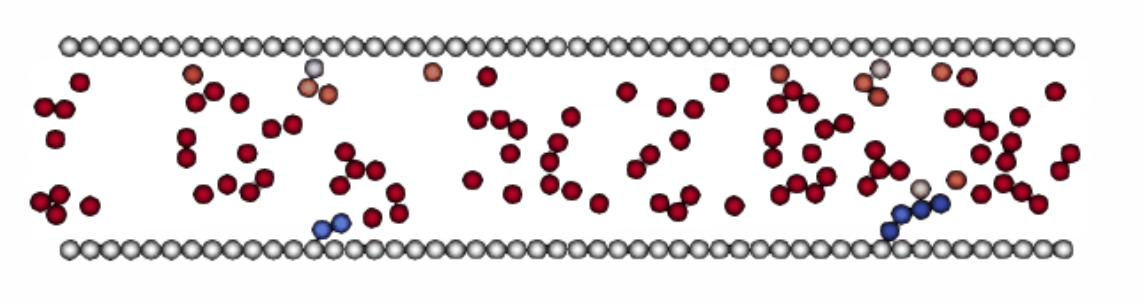
Regards,
Russell
Holt
Email: S352...@student.rmit.edu.au
Erik Navarro
--
You received this message because you are subscribed to the Google Groups "hoomd-users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to hoomd-users...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/hoomd-users/d25aed46-483c-4734-9df4-c0a356773f95n%40googlegroups.com.
Erik Navarro
Russell Holt
Thank you for the suggestion of using the group command, unfortunately I will need to include the wall particles in the integrator in order to correctly build the hydrodynamic coupling tensor. Including the stationary particles is what allows for the resistance tensor to be built which calculates the forces/velocities of the flowing particles, thus I need some way to hold these particles in place irregardless of forces applied. I was hoping there may be a functionality to zero out forces, or maybe set an infinite mass to avoid movement.
Joshua Anderson
In HOOMD-blue, the integration method is responsible for applying the equations of motion to all particles in the given group. Since the integration method you are using is provided by a plugin, you need to ask the plugin authors for support with this feature request.
------
Joshua A. Anderson, Ph.D.
Research Area Specialist, Chemical Engineering, University of Michigan
> On Nov 29, 2021, at 6:37 AM, Russell Holt <rho...@gmail.com> wrote:
>
> Hi Erik
>
> Thank you for the suggestion of using the group command, unfortunately I will need to include the wall particles in the integrator in order to correctly build the hydrodynamic coupling tensor. Including the stationary particles is what allows for the resistance tensor to be built which calculates the forces/velocities of the flowing particles, thus I need some way to hold these particles in place irregardless of forces applied. I was hoping there may be a functionality to zero out forces, or maybe set an infinite mass to avoid movement.
>
> Regards,
> Russell
>
> On Monday, November 29, 2021 at 10:13:29 AM UTC+11 erik.j....@gmail.com wrote:
> Also, in the case of brownian dynamics (which is what I know best), simply omitting a group from an integrator will hold the particles fixed. Interparticle interactions (such as leonard jones) can still be calculated. Perhaps you can do something similar with your simulations. I think hoomd.rigid is really intended to hold particles together relative to one another while allowing the entire rigid body to move.
>
> On Sun, Nov 28, 2021 at 3:30 AM Russell Holt <rho...@gmail.com> wrote:
> Hi all
>
> I am new to HOOMD-Blue however have some experience in other MD packages (LIGGGHTS/LAMMPS). I have recently installed the Fast Stokesian Dynamics (FSD) package ‘PSEv3’ and am interested in applying this package to a pressure driven channel flow.
>
> I have run the included test simulation which comes with the package which uses shearing walls to imply a fluid flow, however what I would like to do is a little different and I’m not sure if this is something HOOMD can do.
>
> I would like to have fixed particles which make up the walls of the channel, and then have a separate flowing group. I believe I should be able to use the ‘rigid’ constraint on the wall particles and then apply the PSEv3 to resolve the hydrodynamic interactions, (with Stokesian Dynamics, the fixed wall particles are required to be included in a mobility matrix which describes the flow of the particles moving through the channel, thus the wall needs to be particles). My problem at the moment is that I cannot see how to make these separate groups for the wall particles. Can the init.read_getar command be used to read in a set of particles, fix these and then create a lattice of particles for the flowing group?
>
> A simple 2D example of this system being run in LAMMPS can be seen attached, where the white particles on the top and bottom are effectively ‘frozen’, still allowing interactions to be calculated, however themselves not moving. This will create the desired parabolic poiseulle velocity profile when a pressure gradient is applied across all particles.
>
>
>
>
>
> Russell Holt
> Email: S352...@student.rmit.edu.au
>
>
>
> --
> You received this message because you are subscribed to the Google Groups "hoomd-users" group.
> To unsubscribe from this group and stop receiving emails from it, send an email to hoomd-users...@googlegroups.com.
> To view this discussion on the web visit https://groups.google.com/d/msgid/hoomd-users/d25aed46-483c-4734-9df4-c0a356773f95n%40googlegroups.com.
>
> --
> You received this message because you are subscribed to the Google Groups "hoomd-users" group.
> To unsubscribe from this group and stop receiving emails from it, send an email to hoomd-users...@googlegroups.com.
Michael Howard
Russell Holt
Thank you both for your help, I have been successful in implementing both azplugins & PSEv3 and am getting some promising results!
I do have one question which I can't seem to find the answer for and was hoping to clarify if this is likely an issue with the plugin, hardware or something else
Currently when I run the simulation I am getting inconsistent segmentation faults (See below image) at different times. This will occur without changing any inputs and will happen at a different step each time.
This may be due to the stochastic nature of the integration method, but I thought it best to see if this is an error anyone has experience with and can point me in the right direction to solve it?
I am not sure if this would have an impact, but I am running HOOMD on WSL2 currently.
Michael Howard
tom...@umich.edu
Russell Holt
It appears that I do not get these errors when using the BD integrator with the same simulation setup in all other regards.
Upon further investigation it seems that this error only occurs when the flowing particles are too close to the restrained wall particles, this leads me to the assumption that the PSE integrator may not be able to correctly integrate the restrained particles, but I will need to confirm with the plugin authors.
Hi Tom
Thank you for that link and feedback, I guess I will need to run some tests in future on a linux system as my simulations will be using the GPU. In your experience is there anything that can be done to improve these issues? And do you find these errors are consistent across specific integrators or completely random?
tom...@umich.edu
Michael Howard
Russell Holt
I have compiled HOOMD as single precision which was what was specified in the PSEv3 plugin instructions, I can try recompiling as double precision if that has any chance of helping?
May I ask if there is any way to dump additional logs from HOOMD? I am currently dumping the configuration of the particles as a GSD file which is giving some insight (perhaps alluding to an incorrect setup), however I cannot use a debugger (I believe? I am new to this!) since I am running the simulation on WSL2.
In regards to a possible incorrect setup, I do believe that using the restraints on the particles may unfortunately be the incorrect approach based on previous stokesian dynamics papers where the wall particles were set to a constant 0 velocity (Ref Page 162: https://doi.org/10.1017/S0022112094002326), the forces from the wall particles appears to be imposing an incorrect force back into the flowing particle.
Without knowing the ramifications of doing this within HOOMD, is it possible to make a function which will hold a constant velocity of zero for a group of particles? something similar to "hoomd.md.force.constant". If this is possible can anyone point me to the best example function/plugin to use as a basis to build this?
Thank you all for all your help!
