Help interpreting GSEA results
Thorben Sauer
Castanza, Anthony
Hi Thorben,
Unfortunately it appears as if the full supplementary data for this paper is no longer available, however, a selection of qPCR validated genes was available in the manuscript text, I was able to confirm that their top hits KARS (now KARS1), AARS (now AARS1), and TARS (now TARS1) are present as expected in the PROVENZANI_METASTASIS_UP signature – so, our signature annotation does appear to match was was presented in the original publication – at least, as much of it as is currently accessible.
Original Publication (note KARS)
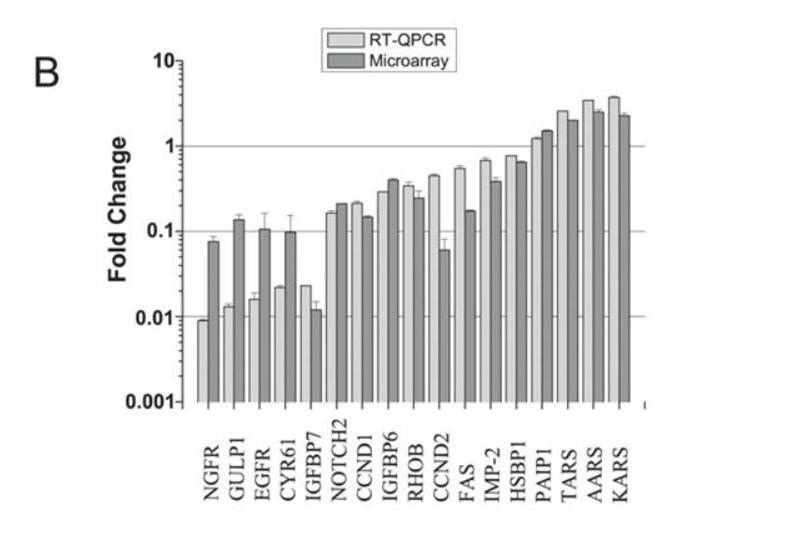
However, looking deeper at the GEO data, I do, in fact, see what you see here:
GEO Data (KARS, same probe as MSigDB UP Signature):

I would have been inclined to suggest that this was an error in the submission to GEO where the annotation was inadvertently switched, but in light of your additional analysis I think the issue here might be slightly more insidious – that the phenotypes were inadvertently switched in the original published analysis.
Thank you for bringing this to our attention. I’m CC’ing the author of the original publication here (Alessandro Provenzani), perhaps they have some additional insight on what might have happened here and what, if any, corrections might be possible.
-Anthony
Anthony S. Castanza, PhD
Department of Medicine
University of California, San Diego
--
You received this message because you are subscribed to the Google Groups "gsea-help" group.
To unsubscribe from this group and stop receiving emails from it, send an email to gsea-help+...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/gsea-help/721575c0-ad13-4929-94ec-6cbf7fec91e2n%40googlegroups.com.
Castanza, Anthony
Hi Thorben,
Unfortunately it appears as if the full supplementary data for this paper is no longer available, however, a selection of qPCR validated genes was available in the manuscript text, I was able to confirm that their top hits KARS (now KARS1), AARS (now AARS1), and TARS (now TARS1) are present as expected in the PROVENZANI_METASTASIS_UP signature – so, our signature annotation does appear to match was was presented in the original publication – at least, as much of it as is currently accessible.
Original Publication (note KARS)
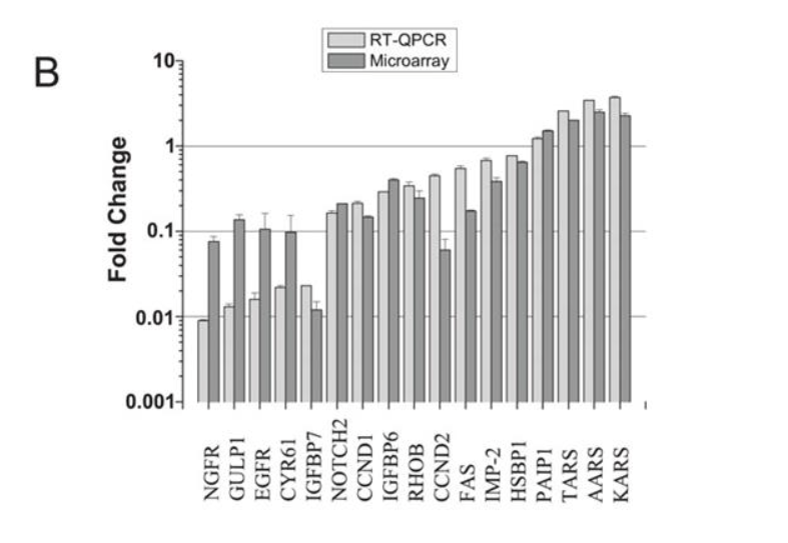
However, looking deeper at the GEO data, I do, in fact, see what you see here:
GEO Data (KARS, same probe as MSigDB UP Signature):

I would have been inclined to suggest that this was an error in the submission to GEO where the annotation was inadvertently switched, but in light of your additional analysis I think the issue here might be slightly more insidious – that the phenotypes were inadvertently switched in the original published analysis.
Thank you for bringing this to our attention. I’m CC’ing the author of the original publication here (Alessandro Provenzani), perhaps they have some additional insight on what might have happened here and what, if any, corrections might be possible.
-Anthony
Anthony S. Castanza, PhD
Department of Medicine
University of California, San Diego
(Resent due to address failure)
Thorben Sauer
Thorben Sauer
Castanza, Anthony
Hi Thorben
No, I haven’t heard anything back from the author. If we’re unable to get a clarification, I’m currently inclined to withdraw the set however we haven’t made a final determination yet. It may be possible to rework the set from just the GEO data in a satisfactory way.
-Anthony
Anthony S. Castanza, PhD
Department of Medicine
University of California, San Diego
To view this discussion on the web visit https://groups.google.com/d/msgid/gsea-help/311ee0c2-4729-4195-99c1-ea1a3d0b2acan%40googlegroups.com.
