I am having difficulty understanding how to make use of the landmarks created through the function digit.curves + a question about importing outlines.
1,057 views
Skip to first unread message
Mykolas Imbrasas
Oct 16, 2017, 7:48:29 PM10/16/17
to geomorph R package
Hello there,
My question is about how to use the equidistant semilandmarks that are generated by Geomorph from imported X and Y coordinates of an outline. I am attaching an image with a somewhat crude representation of the landmarks that I wish to place on a specimen. The red dots represent fixed landmarks, which I successfully managed to place via the digitize2d function and used for a GP and PC analyses. The blue dots represent the semilandmarks that I want to place.
Reading the guide there are, to my knowledge, two ways to place semilandmarks on 2D images. Either manually in digitize2d and then use the defines.sliders function when conducting a gpa, or use the digit.curve function which places equidistant semilandmarks along the outline of an object based on the imported X and Y coordinates of an outline.
I prefer the latter method, because it is much less time consuming. However, is there a way to include or "attach" these Geomorph generated smilandmarks to a specimen with digitized fixed landmarks? Is there a way to save them? And how do I actually make use of them if I have more than 100 specimens and want to assign a set of these equidistant semilandmarks for each specimen? Or in other words, can I somehow attach these semilandmarks to a specimens in a TPS file which contains fixed landmarks and have Geomorph automatically treat them as sliding landmarks in the GP analysis, or use the define.sliders function to do that?
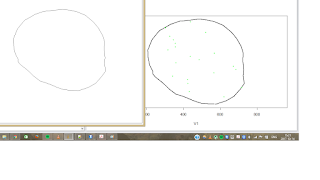
The other thing that I am interested about st the extraction of X and Y coordinates from an outline of an image in ImageJ. I used the procedure as described in the guide but when I upload the X and Y coordinates to R and use the digit.curve function this is the result I get:
I am not sure what is causing it, but I managed to circumvent this issue by using the following procedure:
Image -> Type -> 8 bit.
Process -> Find Edges.
Image -> Adjust -> Threshold. Both parameters set to 255.
Process -> Binary -> Skeletonize.
Then I use the wand tool to select the outline and save the X and Y coordinates. But when I upload the coordinates to R and plot them the image becomes flipped and the outline has a broken line in some places. However, the digit.curves function works WELL:
To fix this issue, here some code that some good internet Samaritan introduced me to:
h = getHeight();
run( "Make Binary" );
run( "Skeletonize" );
run( "Invert LUT" );
doWand( 0, 0.5*h );
getSelectionCoordinates( x, y );
tabDelText = "";
for ( i = 0; i < x.length; i++ ) {
tabDelText += "" + x[i] + "\t" + h - y[i] + "\n";
}
path = getDirectory( "Choose a Directory" );
File.saveString( tabDelText, path + "myTabDelText.txt" );
The highlighted portion of the code tells ImageJ to flip the Y coordinate system, because apparently they start at a different end in R.
What is causing the broken line in the outline that I upload using the method described above? R and Geomorph clearly recognize the
non displayed coordinates in these broken lines.
Best,
Mykolas.
Mykolas Imbrasas
Oct 16, 2017, 9:05:20 PM10/16/17
to geomorph-...@googlegroups.com
P.S.
Sorry for the typos and grammatical errors. I was too quick to post the topic when I should have devoted a little bit more time to proof-reading it.
--
You received this message because you are subscribed to a topic in the Google Groups "geomorph R package" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/geomorph-r-package/WXPQPrIsWx4/unsubscribe.
To unsubscribe from this group and all its topics, send an email to geomorph-r-package+unsub...@googlegroups.com.
To post to this group, send email to geomorph-r-package@googlegroups.com.
Visit this group at https://groups.google.com/group/geomorph-r-package.
To view this discussion on the web, visit https://groups.google.com/d/msgid/geomorph-r-package/9112e5e4-9d25-478e-b838-83351666a46e%40googlegroups.com.
For more options, visit https://groups.google.com/d/optout.
Emma Sherratt
Oct 17, 2017, 1:36:43 AM10/17/17
to geomorph R package
Dear Mykolas,
Semilandmarks along a curve can be digitised in a variety of ways. You have done it one way, the "manual" way, whereby you by eye place semilandmarks on a curve using digitize2d (but you can achieve the same using any other digitizing software like ImageJ or TPSDIG2). There are several "automatic" ways: One is to import into R the curve represented as pixel coordinates (using outline and skeletonize kind of functions with ImageJ, saved as a text file and read in as a matrix) and use digit.curves to resample those pixels into equally distantly space semilandmarks. Or you can draw out the curve by hand with the curve pen tool in TPSDIG2 and it will resample the curve into semilandmarks (both of these use the same maths since I took Jim Rohlf's code to write digit.curve and extend into 3D).
In any of these case, if you are dealing with semilandmarks they must be slid during Procrustes superimposition by specifying the curves= option.
For you, the automatic option is the way as you have started to do. For completeness for anyone reading this, there are two further options that are not with TPSDIG.
1) make a set of 100 text files with the x y coordinates of the pixels of the outlines using imageJ (I provide below some ImageJ macro code I wrote to help you batch this and it works for my tadpole datasets). Read these in and use digit.curves on each.
2) Use R package momocs and the function import_Conte, imort_jpg etc. to make the curves (bypasses the imageJ step above) and then use digit.curves on those.
Now the imageJ issue you have is one I have come up against many times. ImageJ flips the y-axis by default, go to: Analyze > Set Measurements > Check Invert Y coordinates. That should be the simple fix
Emma
IMAGEJ Macro Code to Batch convert and save outlines as text files
// Batch Convert
//
// This macro convert all the files in a folder to a text image.
// First it makes the image binary.
// Then it fills the holes.
// Then saves as text image
// To be used on B&W drawings.
dir = getDirectory("Choose Source Directory ");
list = getFileList(dir);
setBatchMode(true);
for (i=0; i<list.length; i++) {
showProgress(i+1, list.length);
open(dir+list[i]);
setOption("BlackBackground", false);
run("Make Binary");
run("Fill Holes");
run("Text Image...");
saveAs("Text Image", dir+list[i]);
close();
Mykolas Imbrasas
Oct 17, 2017, 9:52:21 AM10/17/17
to geomorph R package
Dear Emma,
Thank you for the comment and the macro code which seems very useful.
My sample consists of specimens from different species and I am interested in comparing the morphological features between them. When I place fixed landmarks with the function digitize2d, Geomorph creates a TPS file which contains the landmark coordinates for each specimen. Is there a way to save the semilandmarks generated by the function digit.curves as a TPS file in the same format as digitize2d does? i.e. number of landmarks, their coordinates, scale, and ID. Or, can I add the coordinates of the semilandmarks generated this way to a TPS file that contains fixed landmarks, so I could then use the define.sliders function.
Mykolas.
Emma Sherratt
Oct 17, 2017, 4:46:31 PM10/17/17
to geomorph-...@googlegroups.com
Yes. Simply combine the two data sets into a single 3D array and use writeland.tps()
Emma
--
You received this message because you are subscribed to the Google Groups "geomorph R package" group.
To unsubscribe from this group and stop receiving emails from it, send an email to geomorph-r-pack...@googlegroups.com.
To post to this group, send email to geomorph-...@googlegroups.com.
Visit this group at https://groups.google.com/group/geomorph-r-package.
To view this discussion on the web, visit https://groups.google.com/d/msgid/geomorph-r-package/955ccd46-28aa-4145-989c-30c906848f38%40googlegroups.com.
For more options, visit https://groups.google.com/d/optout.
--
Sent from my thumbs
Mykolas Imbrasas
Oct 24, 2017, 9:59:44 AM10/24/17
to geomorph-...@googlegroups.com
Hi Emma,
I am afraid I do not understand. I am attaching a TPS file of the fixed landmarks that I digitized in R with the function digitize2d and a text file that contains the equidistant semilandmarks that I digitized with the digit.curves function. The first 56 semilandmarks should belong to the first specimen in the TPS files, the second 56 semilandmarks should belong to the second and so on. Do I just copy paste the landmark coordinates of the semilandmarks into the TPS file with the fixed coordinates, or create a new text file with the combined landmarks, then upload it to R with the function read.table() (?), and then convert the data into a 3D array with arrayspecs(A, p, k)?
Thank you.
Emma Sherratt
Oct 24, 2017, 6:49:14 PM10/24/17
to geomorph R package
Mykolas,
When your semilandmarks are still a 3D array object in R (after running digit.curves), it's simple to combine the two using abind() library (install this):
new.coords <- abind(fixed, semis) # where semis is a 3D array of semilandmarks, e.g. from digit.curves; fixed is a 3D array of fixed landmarks, eg. from TPS file
Or without abind you must turn both into 2D arrays and combine as such:
new.coords <- cbind(two.d.array(fixed), two.d.array(semis))
Then you can use arrayspecs() to make new.coords a 3D array again
Emma
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~Emma Sherratt, PhD.University Research FellowDepartment of Genetics and EvolutionSchool of Biological SciencesThe University of AdelaideAdelaide, SA 5005AUSTRALIAHonorary ResearcherSouth Australia MuseumAdelaide, SA 5005AUSTRALIAemail: emma.s...@gmail.commob: +61 4234 19966Twitter: @DrEmSherrattUA ALLY :: Supporting the diversity of sexuality and gender identity at U Adelaide.
Caecilians are legless amphibians...
__ (\ .-. .-. /_") \\_//^\\_//^\\_// `"` `"` `"`
learn more about them here: www.emmasherratt.com/caecilians
--
You received this message because you are subscribed to the Google Groups "geomorph R package" group.
To unsubscribe from this group and stop receiving emails from it, send an email to geomorph-r-package+unsub...@googlegroups.com.
To post to this group, send email to geomorph-r-package@googlegroups.com.
Visit this group at https://groups.google.com/group/geomorph-r-package.
To view this discussion on the web, visit https://groups.google.com/d/msgid/geomorph-r-package/CAAyoj0_n5%2BZe15r%3D8B7zRFX2tHGvsfjA5pzvrcqXPtZYgpWkLQ%40mail.gmail.com.
Mykolas Imbrasas
Oct 25, 2017, 7:39:09 AM10/25/17
to geomorph-...@googlegroups.com
Emma,
I installed abind but when I try to combine the two I get the following error:
Error in abind(fixed, lmks1) :
arg 'X2' has dims=56, 2, 1; but need dims=10, 2, X
I thought that the issue might be n, but when I attempt to merge one set of semilandmarks with the fixed landmarks of one specimen I get the same error. So I think it's the difference in the number of landmarks between the two that causes the issue.
I tried turning them into 2D arrays, however I get the following error when trying to turn the semilandmakrs into a 2D array:
Error in aperm.default(A, c(3, 2, 1)) :
'perm' is of wrong length 3 (!= 2)
I am an absolute beginner in R so this is baffling to me.
Thank you.
Mykolas.
--
To unsubscribe from this group and stop receiving emails from it, send an email to geomorph-r-package+unsubscribe@googlegroups.com.
To post to this group, send email to geomorph-r-package@googlegroups.com.
Visit this group at https://groups.google.com/group/geomorph-r-package.
To view this discussion on the web, visit https://groups.google.com/d/msgid/geomorph-r-package/CAAyoj0_n5%2BZe15r%3D8B7zRFX2tHGvsfjA5pzvrcqXPtZYgpWkLQ%40mail.gmail.com.
--
You received this message because you are subscribed to a topic in the Google Groups "geomorph R package" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/geomorph-r-package/WXPQPrIsWx4/unsubscribe.
To unsubscribe from this group and all its topics, send an email to geomorph-r-package+unsub...@googlegroups.com.
To post to this group, send email to geomorph-r-package@googlegroups.com.
Visit this group at https://groups.google.com/group/geomorph-r-package.
To view this discussion on the web, visit https://groups.google.com/d/msgid/geomorph-r-package/CAF1_pPtGr0VgnUqun4Z0xsdZnvEZDZ_q7Q5a4U%3Dhe2sy3RrJcg%40mail.gmail.com.
Reply all
Reply to author
Forward
Message has been deleted
0 new messages