Doc2vec training takes extraordinarily long time
599 views
Skip to first unread message
Sharon Kang
Aug 14, 2022, 12:57:43 AM8/14/22
to Gensim
Hello All,
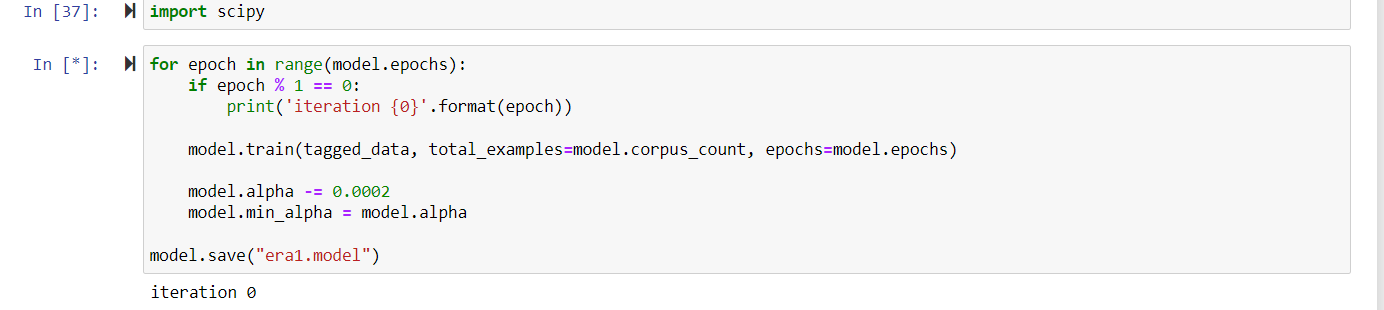
I am currently in the process of training a doc2vec model with about 500,000 datasets.
I imported CSV file by using pandas, using uid column as tags and the text column as words

Tagged Data was created very well, able to print and check it.

I was also able to build the corpus as well and use the get_vecattr or index_to_key methods.

But training the model takes an extraordinarily long time even though the official document of gensim said it shouldn't take more than 2 minutes.. For the model training below, it took more than 8 hours but was iterated only 9 times.

I've been dealing with this problem for more than a week, and I don't see why it takes such a long time.. is there a bug /error in my code?
Please let me know how to fix this problem. Thank you so much!!!
Best wishes,
Sharon Kang
Gordon Mohr
Aug 15, 2022, 3:54:38 PM8/15/22
to Gensim
Regarding reasonable performance expectations:
Essentially, no adequate training data for for `Doc2Vec` will result in training completing in 2 minutes. Only tiny toy-sized demo datasets will complete in that amount of time. Even partial replication of the original `Paragraph Vector` paper IMDB movie-reviews results (~100,000 short texts of no more than a few hundred words each) takes at least tens-of-minutes on common systems. It's been a while since I did a bulk training on a Wikipedia dump with millions of documents, some with thousands of words, but I recall that taking 14h+.
If there's some part of the official docs that sets an unrealistic expectation, please provide a link so it can be corrected.
Specific to what you've shown:
Your data setup & general steps seem proper; however, there is rarely any reason for users to call `.train()` multiple times inside their own loop, and manage the `alpha` value decay themselves. It's unnecessary, overcomplicated, & error-prone – but for some reason this anti-pattern has been widely copied by poor-quality online tutorials that barely seem to understand what they're doing (much less explain it to their learning readers).
More discussion at this SO answer: https://stackoverflow.com/questions/62801052/my-doc2vec-code-after-many-loops-epochs-of-training-isnt-giving-good-results/62801053#62801053
Specifically in your case, 40 of your own loops outside a `.train()` that is itself doing 40 internal epochs means you're doing 1600 total passes over your data – an overkill surely not intended. That's likely the biggest reason for surprisingly lengthy training time.
Further, manually decrementing starting `alpha` from its normal default of 0.025 by 0.0002, 40 times, only brings it down to 0.017 – normal SGD would decay it fully to a value-near-0.0. But because this antipattern code only tampers with `min_alpha` *after* the 1st call, you'll actually have one proper full decay, using the proper defaults automatically (on the 1st loop), then 39 more that are nonsense. This won't lengthen run-time, but it will make results unrrepresentative (& probably bad).
Call `.train()` exactly once, unless you're an expert with a clear idea of why you're doing something very non-standard & error-prone.
If you want to see incremental progress, enabling logging at the INFO level is a good idea. (If you need to log some interim evaluation, it's also possible to set up end-of-epoch callbacks. But keep in mind that an evaluation after, say, 20 epochs of a planned 40 epoch run will be measuring something quite different from a evaluation of a true 20 epoch run at its end.)
- Gordon
Message has been deleted
Sharon Kang
Aug 17, 2022, 8:51:53 AM8/17/22
to gen...@googlegroups.com
Dear Gordon,
** updated the typo**
Thank you for your reply! I erased the for loop and placed the .train method outside of it, and training took about 2 hours!
I wonder why that anti-pattern is widespread.. it made me so confused.
This is a sentence from the official gensim document : (https://radimrehurek.com/gensim/auto_examples/tutorials/run_doc2vec_lee.html) : ` Next, train the model on the corpus. If optimized Gensim (with BLAS library) is being used, this should take no more than 3 seconds. If the BLAS library is not being used, this should take no more than 2 minutes, so use optimized Gensim with BLAS if you value your time`
Now that I think of it, maybe it only implied that 300 small dataset should take no more than 2 minutes but I think it can be misread to be denoted for all datasets.
Furthermore, I am in the process of making a doc2vec trained model for a t-SNE plot ... but want to color the words from a specific category to check how the words from that category are scattered but I wonder if it's possible..
I looked through the replies you left on this group and it seemed like you recommend a single unique tag to one document (obviously category names are neither single nor unique..)
This SO discussion tells that I can ( https://stackoverflow.com/questions/50501364/word2vec-tsne-plot) but I am not super convinced how to since I didn't use category when making a tagged_document.
I have another dataset that just extracted documents from that one certain category.. can I make a t-SNE plot for that dataset and overlap these like layers? would it be possible?
Many thanks,
Sharon
--
You received this message because you are subscribed to a topic in the Google Groups "Gensim" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/gensim/ZnuJkxWT6ps/unsubscribe.
To unsubscribe from this group and all its topics, send an email to gensim+un...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/gensim/b8fbce6c-03cb-469a-9844-46e1ceeb13f6n%40googlegroups.com.
Gordon Mohr
Aug 17, 2022, 2:46:44 PM8/17/22
to Gensim
On Wednesday, August 17, 2022 at 5:51:53 AM UTC-7 kan...@mtholyoke.edu wrote:
Dear Gordon,** updated the typo**Thank you for your reply! I erased the for loop and placed the .train method outside of it, and training took about 2 hours!I wonder why that anti-pattern is widespread.. it made me so confused.
I'ts frustrating, because one early official example of an *atypical* situation where someone might want to do such a loop got widely copied into other tutorials, often erroneously, & it just keeps replicating, with fumbling-through-it-without-real-understanding tutorial authors copying it amongst themselves.
This is a sentence from the official gensim document : (https://radimrehurek.com/gensim/auto_examples/tutorials/run_doc2vec_lee.html) : ` Next, train the model on the corpus. If optimized Gensim (with BLAS library) is being used, this should take no more than 3 seconds. If the BLAS library is not being used, this should take no more than 2 minutes, so use optimized Gensim with BLAS if you value your time`Now that I think of it, maybe it only implied that 300 small dataset should take no more than 2 minutes but I think it can be misread to be denoted for all datasets.
Yes, that comment was only meant to pertain to that tiny toy-sized demo. I've opened an issue to improve the wording: https://github.com/RaRe-Technologies/gensim/pull/3381/files
Furthermore, I am in the process of making a doc2vec trained model for a t-SNE plot ... but want to color the words from a specific category to check how the words from that category are scattered but I wonder if it's possible..I looked through the replies you left on this group and it seemed like you recommend a single unique tag to one document (obviously category names are neither single nor unique..)This SO discussion tells that I can ( https://stackoverflow.com/questions/50501364/word2vec-tsne-plot) but I am not super convinced how to since I didn't use category when making a tagged_document.I have another dataset that just extracted documents from that one certain category.. can I make a t-SNE plot for that dataset and overlap these like layers? would it be possible?
You can always try applying t-SNE - whether its radical dimensionality-reduction shows anything interesting on your data can only be seen by tinkering. Even if you have categories that *weren't* fed to the `Doc2Vec` (or `Word2Vec` etc) training, you should be able to use those external categories later as labels on other plots. Just merge/cross-reference those labels in, from whatever their authoritative source is, rather than expecting them from the `Doc2Vec` model directly.
(Yes, one `tag` per document, as a unique ID, is the original & most-typical use of `Doc2Vec`, such that each document gets a distinct vector. Mixing in other mutliple-tags, perhaps relfecting other known-aspects, can sometimes help but increases complexity and other tradeoffs. As a hint of one class of concerns: if you mix-in a known-label during `Doc2Vec` training, that label will get, via training, a single summary vector. That can be nifty! But in the very-high-dimensionality spaces of documents/records, the true group of documents with that label may be some odd, lumpy, stretched, donut-holed, etc region, that's *not* well summarized by one single crude centroid point. If you use an unsupervised feature-extraction technique like `Doc2Vec` to let each document, by its own terms, land in a unique appropriate place, then *downstream* clustering/classification/etc steps that are fed those coordinates, and the labels, have a chance to learn more-sophisticated representations of the label, not limited to a single summary point.)
- Gordon
Reply all
Reply to author
Forward
0 new messages
